Combination and application of lamp primers for detection of 4 ophthalmic infection viruses
A primer combination and primer set technology, applied in microorganism-based methods, microorganisms, recombinant DNA technology, etc., can solve the problems of long PCR detection time, application limitations, and high false positive rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0169] Embodiment 1, primer design and preparation
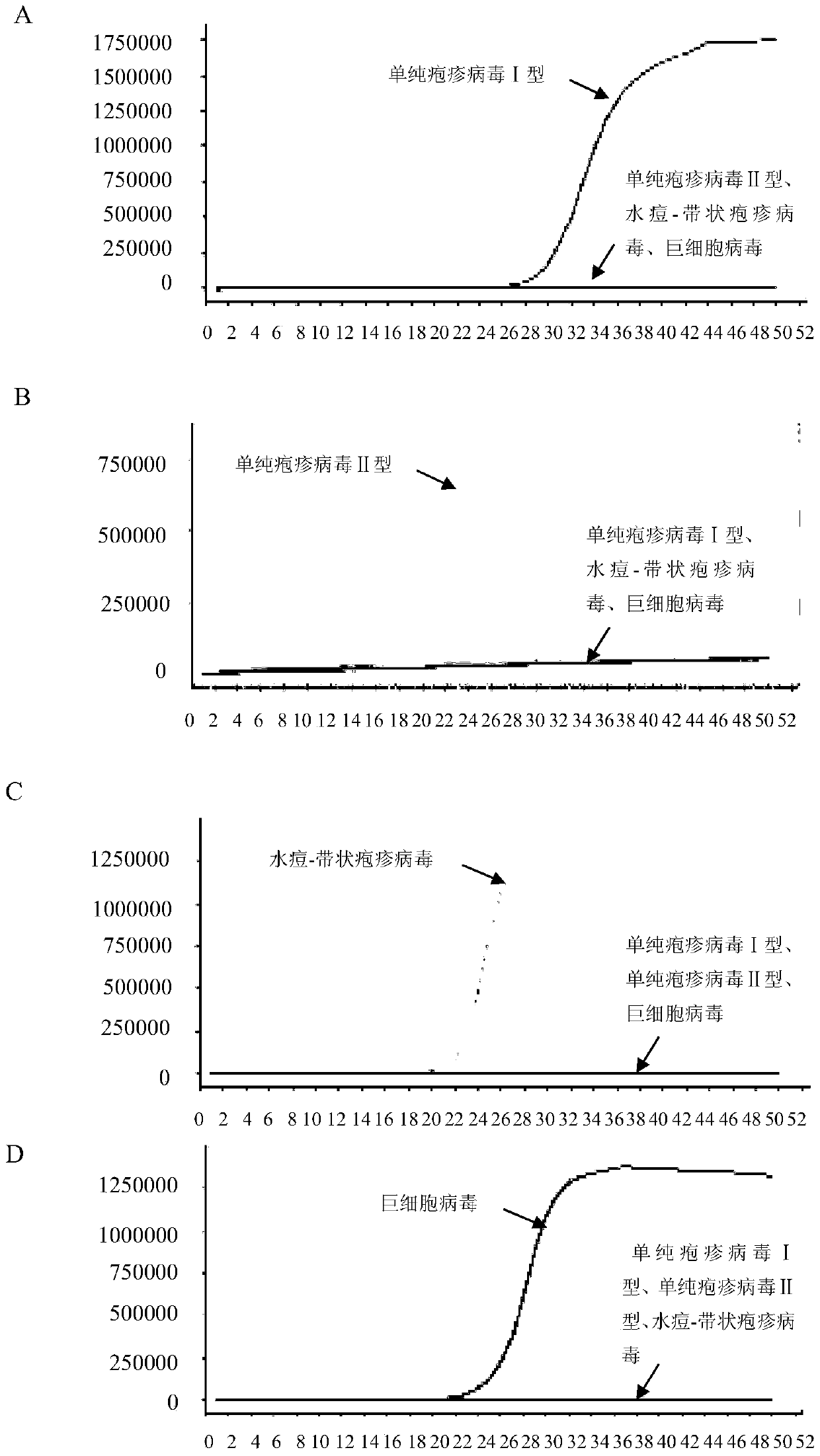
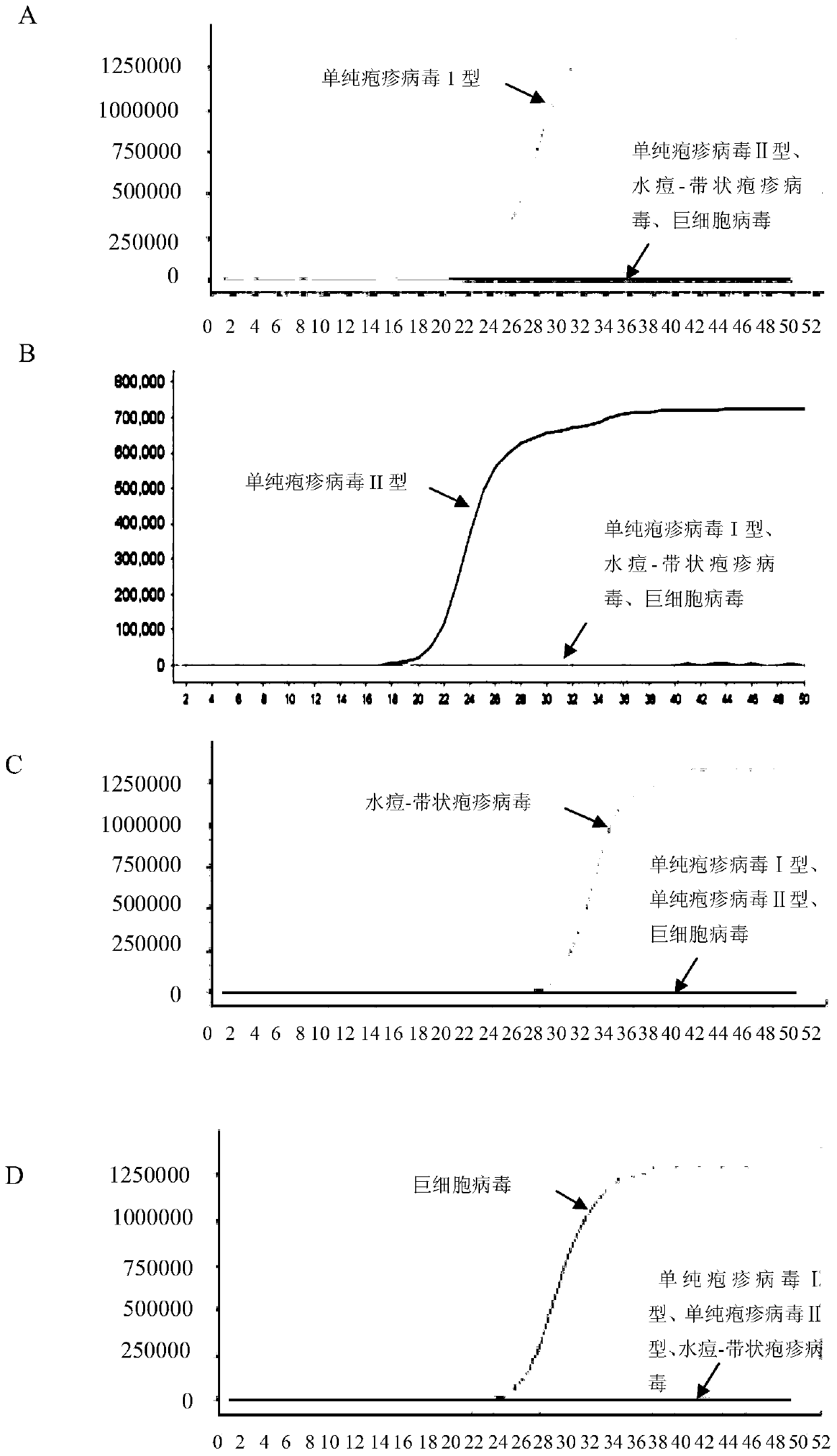
[0170] A large number of sequence analyzes and comparisons were carried out to obtain several primers for identifying herpes simplex virus type I, herpes simplex virus type II, cytomegalovirus and varicella-zoster virus. Preliminary experiments were performed on each primer to compare their sensitivity and specificity, and finally four sets of LAMP primers for identifying HSV-I, HSV-II, varicella-zoster virus and cytomegalovirus were obtained.
[0171] A primer set for identifying herpes simplex virus type I, including 2 outer primers (F3-1, B3-1), 2 inner primers (FIP-1, BIP-1) and 2 loop primers (LF-1, LB-1), each primer sequence is as follows (5'→3'):
[0172] F3-1 (sequence 1 of the sequence listing): TGCGGCTCGTGAAGACC;
[0173] B3-1 (sequence 2 of the sequence listing): TACGGCGACGGTGCGTA;
[0174] FIP-1 (SEQ ID NO: 3 of the SEQUENCE LISTING): TACTTACAGGAGCCCTTGGGACTGGACGGAGATTACACAGCG;
[0175] BIP-1 (SEQ ID NO: 4 o...
Embodiment 2
[0204] Embodiment 2, detection method establishment
[0205] 1. Extract the genomic DNA of the virus to be tested or extract the total DNA of the biological sample to be tested.
[0206] 2. Take the DNA obtained in step 1 as a template, and use the primer set I prepared in Example 1 to perform LAMP amplification.
[0207] Reaction system for LAMP amplification: 1μL 10×ThermoPol Buffer, 1.6μL 5M betaine, 0.1μL 50mg / ml BSA, 0.4μL 100mM MgSO 4 , 0.3 μL 20×EvaGreen, 0.15 μL 100 mM dNTPs, 0.4 μL 8U / ml Bst DNA polymerase large fragment, 1 μL primer mix (F3-1, B3-1, FIP-1, BIP-1, LF-1 and LB-1 mixture), 2ng template DNA, with ddH 2 O to make up to 10 μL. The concentration of each primer in the reaction system is as follows: 0.5 μM F3-1, 0.5 μM MB3-1, 2 μM FIP-1, 2 μM BIP-1, 1 μM LF-1, 1 μM LB-1.
[0208] Reaction program for LAMP amplification: constant temperature at 65°C for 50min. During the reaction process, the fluorescent signal was detected by a fluorescent PCR instrument...
Embodiment 3
[0220] Embodiment 3, reference plasmid construction
[0221] Herpes simplex virus type I reference plasmid: insert the double-stranded DNA molecule shown in sequence 25 of the sequence listing between the ApaI and SacI restriction sites of the pGEM-TEasy Vector vector to obtain the reference plasmid.
[0222] Herpes simplex virus type II reference plasmid: insert the double-stranded DNA molecule shown in sequence 26 of the sequence listing between the ApaI and SacI restriction sites of the pGEM-T Easy Vector vector to obtain the reference plasmid.
[0223] Varicella-zoster virus reference plasmid: insert the double-stranded DNA molecule shown in sequence 27 of the sequence listing between the ApaI and SacI restriction sites of the pGEM-T Easy Vector vector to obtain the reference plasmid.
[0224] Cytomegalovirus reference plasmid: Insert the double-stranded DNA molecule shown in sequence 28 of the sequence listing between the ApaI and SacI restriction sites of the pGEM-T Easy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com