A method for constructing wild rice chromosome segment replacement line
A technology of chromosome fragments and replacement lines, which is applied in the field of construction of wild rice chromosome fragment replacement lines, can solve the problems of too large imported fragments, low efficiency of gene cloning, insufficient research and analysis, etc., and achieve the effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1, Identification and Screening of Polymorphic SSR Molecular Markers
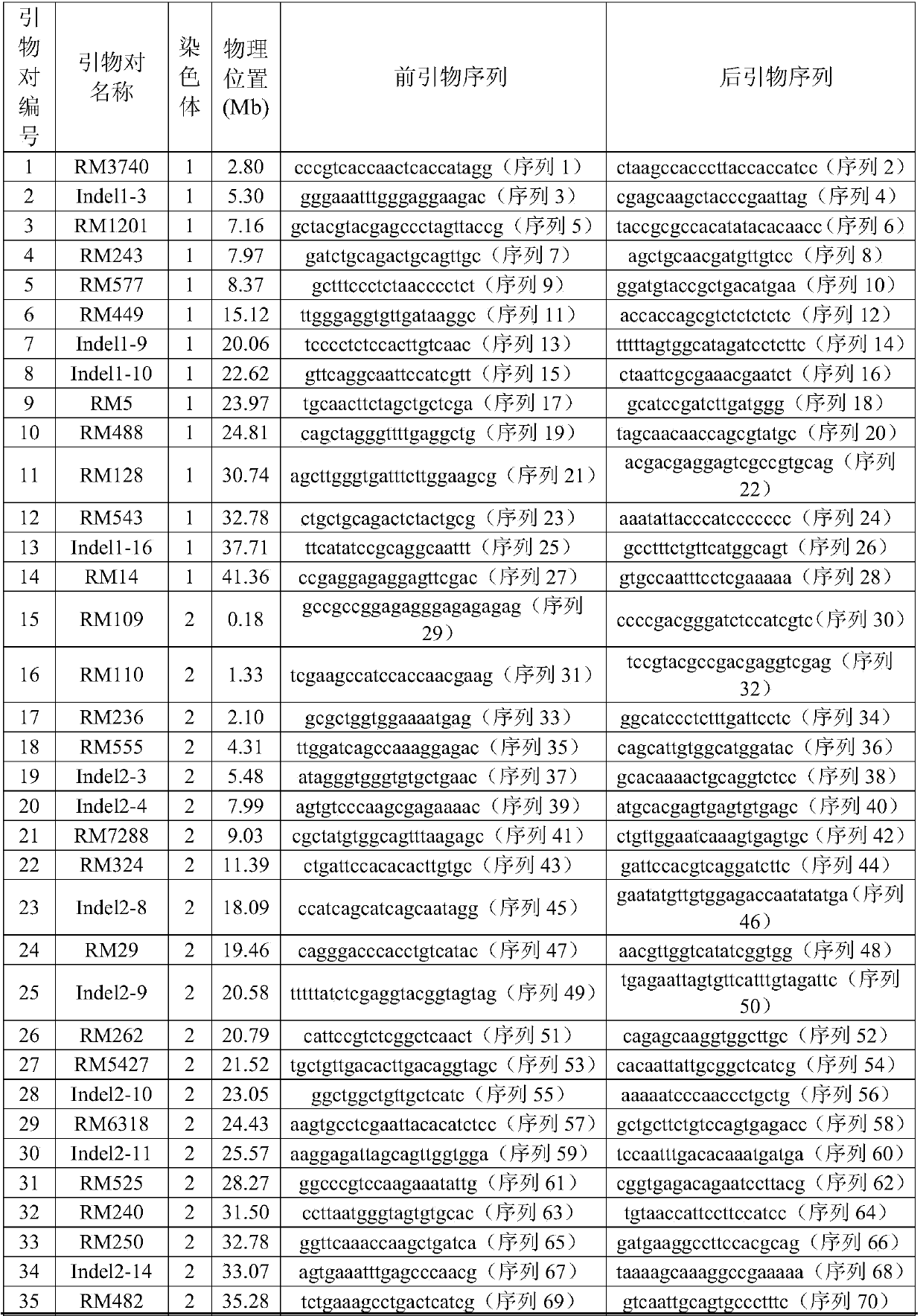
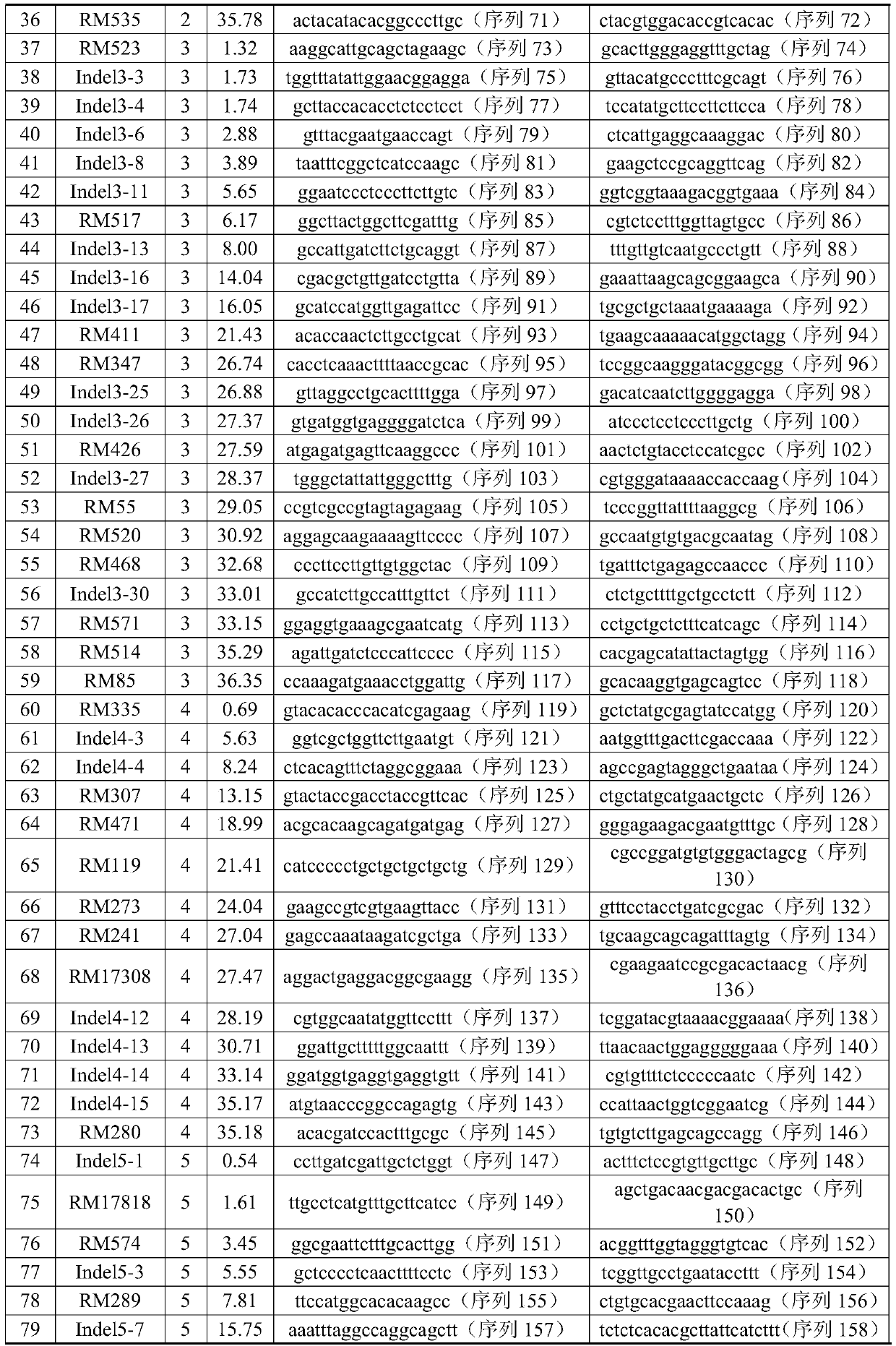
[0062] 1. Select 780 pairs of SSR primers and 175 pairs of InDel primers distributed on 12 chromosomes to screen the primers used to construct the replacement line. The genetic distance of all the primers (unit: cM centimolar root) comes from the website http: / / www .gramene.org.
[0063] 2. 369 pairs of SSR primers were detected in the reference "Eshed Y, Zamir D: A genomic library of Lycopersiconpennellii in L.esculentum: a tool for fine mapping of genes. Euphytica, 1994, 79(3): 175-179." There was polymorphism between the parents (wild rice and cultivated rice 9311), and finally 119 pairs of SSR primers and 62 pairs of InDel primers evenly distributed on 12 chromosomes were selected to construct the replacement line. The information and sequences of the 181 pairs of primers are shown in Table 1, and the average genetic distance between each pair of primers is 5.7cM.
[0064] Table 1. Info...
Embodiment 2
[0070] Embodiment 2, the construction method of wild rice chromosome segment replacement line
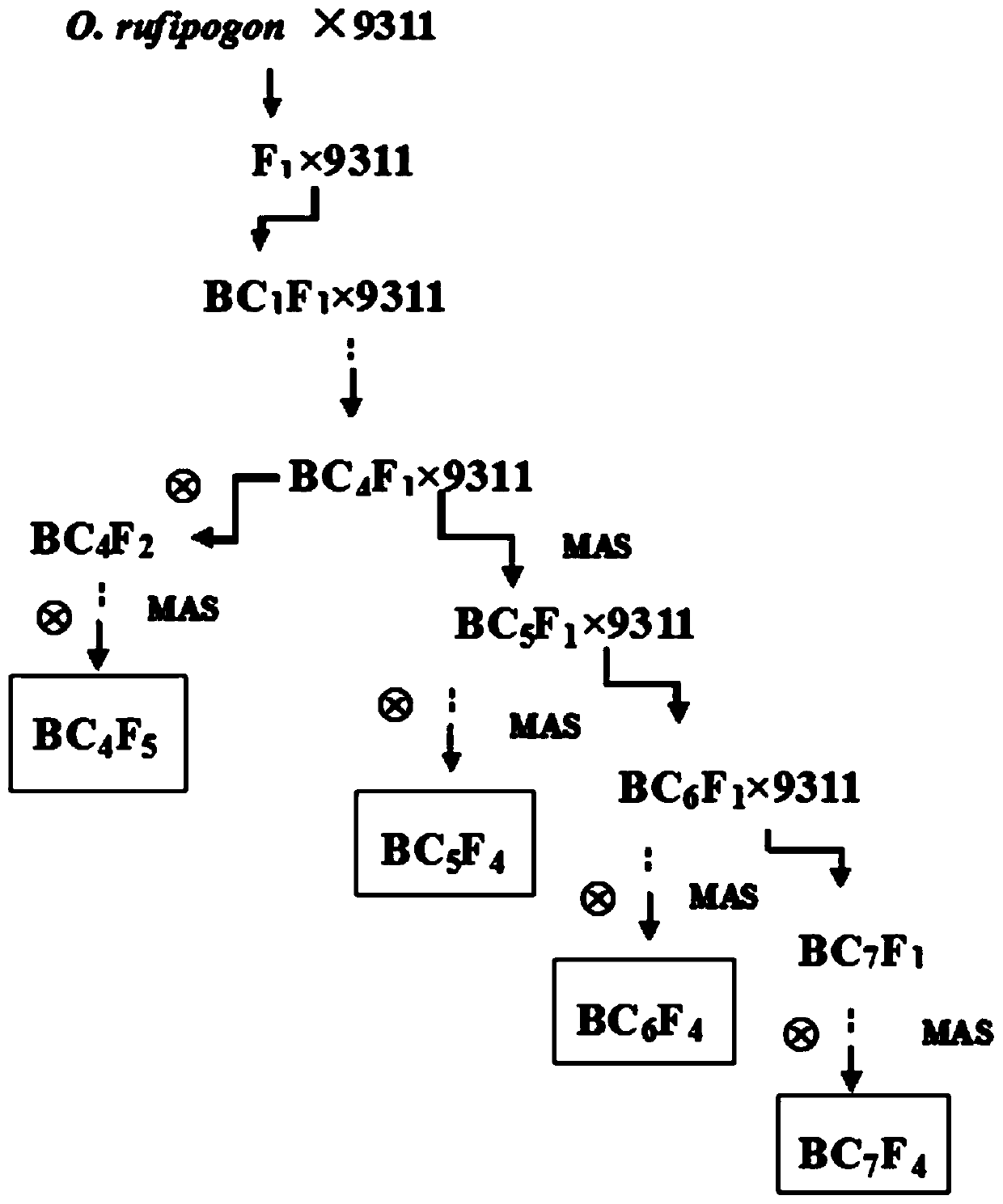
[0071] The method for constructing the wild rice chromosome segment replacement line of the present invention is to use continuous backcrossing combined with MAS to detect molecular markers from the BC3 generation, perform selective backcrossing according to the distribution of exogenous DNA fragments, and continuously track and detect up to the high-generation backcrossed offspring , to establish a chromosome segment replacement line of the whole genome series of wild rice. The construction flow diagram of the wild rice chromosome segment replacement line of the present invention is as follows figure 1 As shown, the construction method of the wild rice chromosome segment replacement line of the present invention is specifically as follows:
[0072] 1. Using the cultivated rice 9311 as the female parent and the common wild rice as the male parent, carry out hybridization to obtain ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com