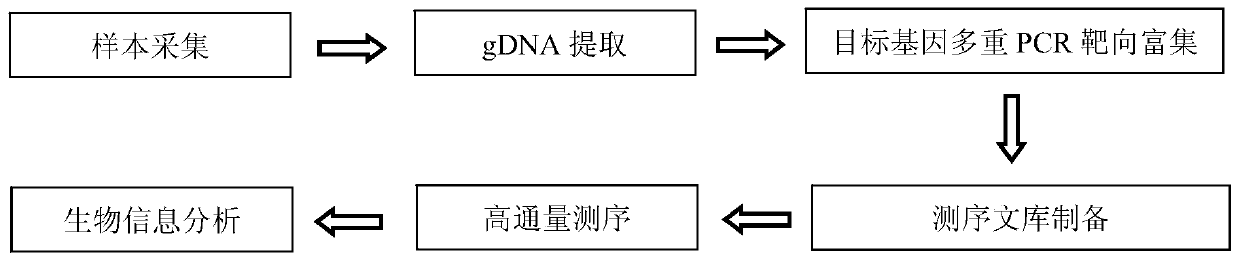
A kind of MMR gene mutation detection kit
A kit and gene mutation technology, applied in the fields of molecular biology and medicine, can solve problems such as expensive, time-consuming sequencing, and inability to meet detection needs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0115] Example 1 Primer Design and Synthesis
[0116] The present invention designs 87 pairs of Primers for a total of 11419 bases in 73 exons of 5 genes (MSH1, MLH2, MSH6, PMS1, PMS2), and the length of the PCR amplification product is 257-528bp. The combined probe primer can Covering all exon regions of 5 MMR genes (see Table 1). Primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd. See Table 2 for primer sequence information.
[0117] Table 1: Reference sequence information table for primer design
[0118]
[0119] Table 2: 87 pairs of primers targeting 5 MMR genes
[0120]
[0121]
[0122]
[0123]
Embodiment 2
[0124] Example 2 Primer combination and multiplex PCR targeted enrichment method
[0125] 1. Experimental materials
[0126] (1), the main reagents
[0127] 1. Fluorescence quantitative detection kit: Qubit TM dsDNA HS Assay Kit (500 assays), Lifetechnologies;
[0128] 2. Quantitative detection kit: Agilent DNA 7500 and 12000Kit, Agilent Company, the kit includes;
[0129] 3. Multiplex PCR kit: MixMultiplex PCR 5×Master Mix, NEB Company;
[0130] 4. Sequencing library ligation adapter kit: NEXTflex TM DNA Barcode-48, BIOO SCIENTIFIC;
[0131] 5. Sequencing library construction kit: NEXTflex TM Rapid DNA-Seq Kit, BIOO SCIENTIFIC;
[0132] 6. Sequencing kit: MiSeq Reagent Kit v3 (600-cycles), Illumina
[0133] 7. Purification kit: Agencourt AMPure XP beads, Beckman Coulter (Agencourt) company
[0134] (2) Main instruments
[0135] 1. 0.5ul~2ml pipette, Eppendof, Germany;
[0136] 2. Micro centrifuge, Hangzhou Aosheng Instrument Co., Ltd.;
[0137] 3. High-speed ce...
Embodiment 3
[0249] Example 3 Detection of MMR gene mutation in 17 cases of colorectal cancer and 14 normal persons
[0250] One, experimental material, with embodiment 2.
[0251] 2. Sample testing
[0252] (1) Multiplex PCR targeted enrichment
[0253] Genomic DNA was extracted from 31 samples (17 cases of colorectal cancer and 14 cases of normal people) using Qiagen's kit and according to the method in the kit instructions. The genomic DNA of 31 samples was amplified by multiplex PCR using 11 sets of primer pairs as shown in Table 3 of Example 2, and the specific method was the same as in Example 2.
[0254] (2), multiple PCR product purification, specific method is the same as embodiment 2.
[0255] (3), sequencing library construction, specific method is the same as embodiment 2.
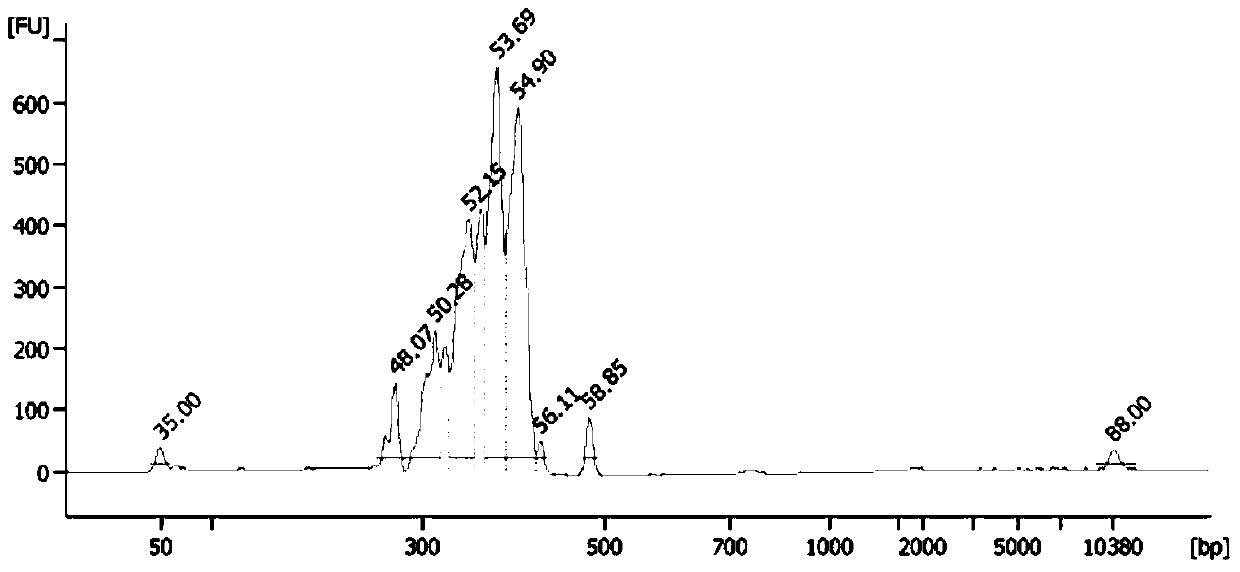
[0256] (4), the quality inspection of the sequencing library, the specific method is the same as that in Example 2.
[0257] 3. High-throughput sequencing, the specific method is the same as that in Ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com