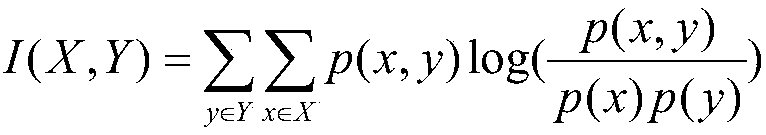A method for screening tumor protein markers based on multi-layer complex network
A complex network and protein technology, applied in the field of screening tumor protein markers based on multi-layer complex networks, can solve problems such as failure to learn, difficult to interpret output results, and long learning time for artificial neural network algorithms, and the method is simple and accurate. high degree of effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
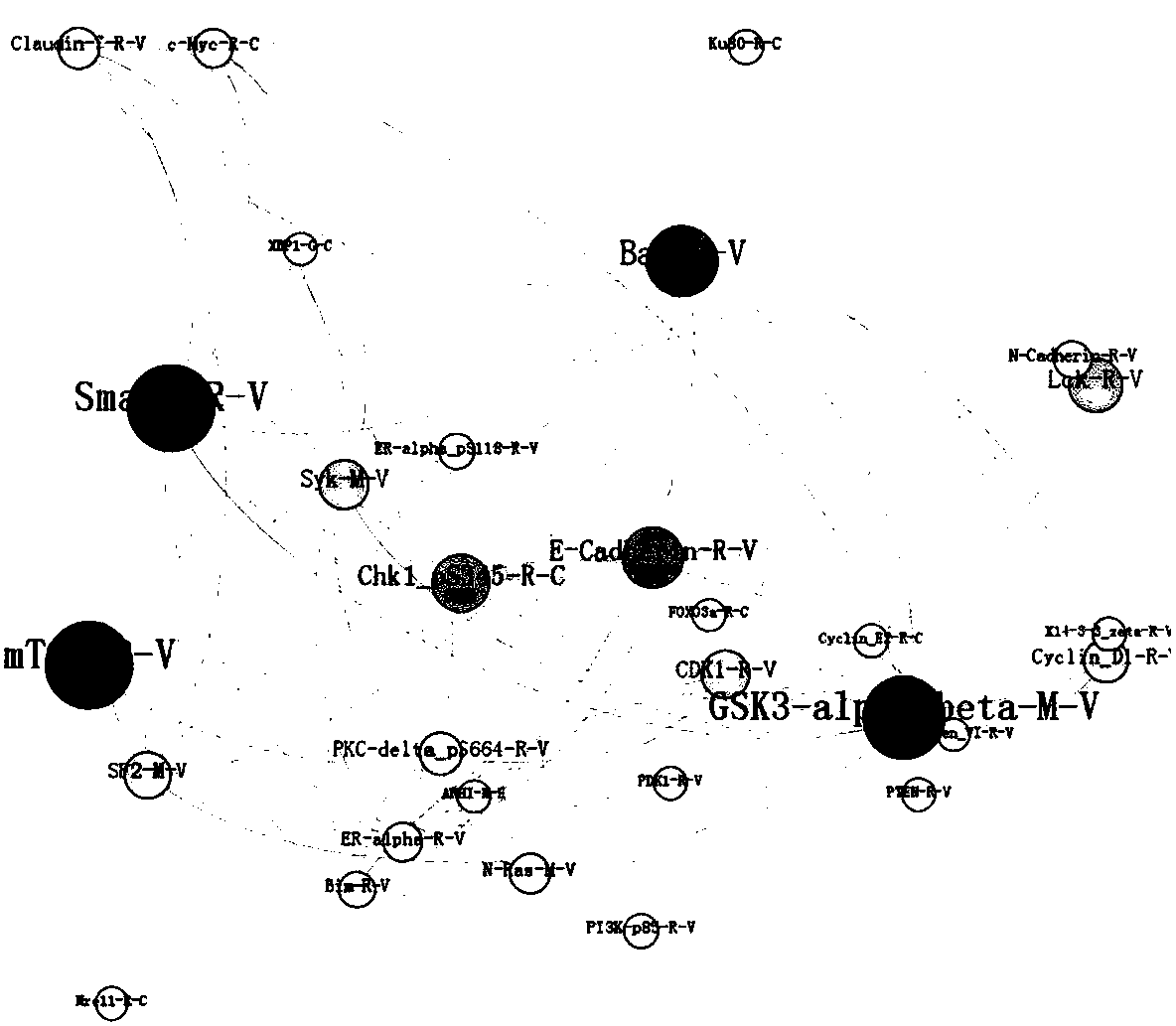
[0067] The source of the research data is The Cancer Genome Atlas / TCGA (https: / / tcga-data.nci.nih.gov / tcga / dataAccessMatrix.htm). Select the data of invasive breast cancer patient protein Experssion-Protein data level 3 to download. Among them, there are 285 protein data, which come from 937 patients. Among the protein expression data, 45 are normal tissue protein data of breast cancer patients, and the rest are tumor tissue protein data of breast cancer patients. In the protein data of normal tissue and tumor tissue, there are many proteins that are not expressed or the expression rate is low. Remove the individuals that do not express the protein, and obtain the protein data of normal tissue and breast tumor tissue with a size of 137×45, that is, select 137 out of 45 patients. Different types of normal tissue protein data and 137 tumor tissue protein data of the same type as normal tissue.
[0068] The random forest model is used to encapsulate and filter the protein data ...
Embodiment 2
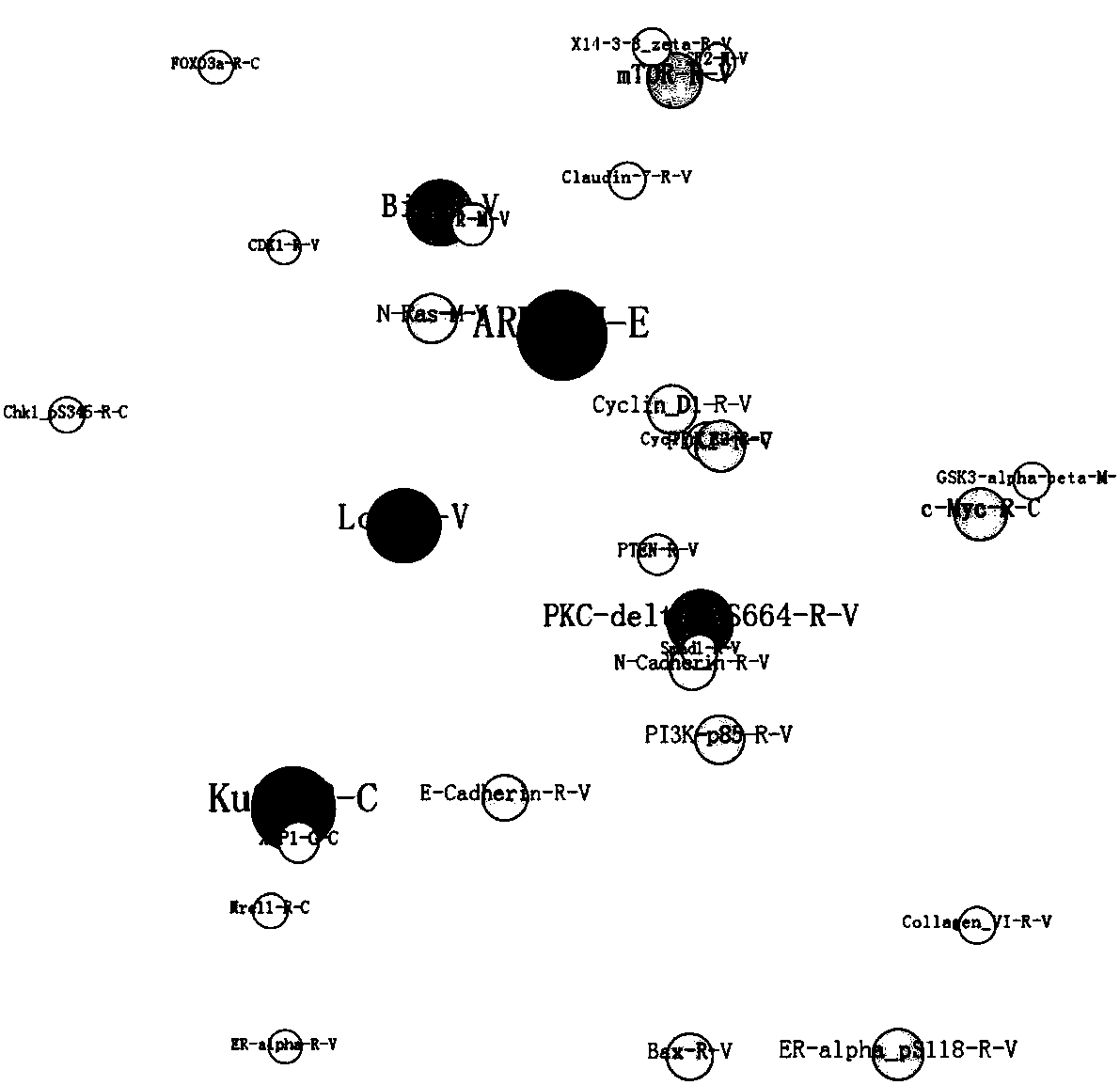
[0077] The protein data of lung cancer patients were downloaded by using the method of Example 1, wherein the protein data were 276 from 166 patients. After deleting the censored data, 137×166 lung cancer tumor tissue protein data are obtained. Since lung cancer patients lack normal tissue data, here we select the normal tissue data of breast cancer patients as a control, that is, select 131 tumor tissue proteins of the same type from 166 patients data and 131 normal tissue protein data from 45 patients.
[0078] The random forest model is used to encapsulate and filter the protein data of normal tissue and tumor tissue of lung cancer patients to select the best subset. In order to select the protein subset with the smallest number of genes and maintain the highest classification accuracy, a ten-fold cross-validation test was used to evaluate the classifier model, and the protein classification results are shown in Table 4. For the breast cancer data set, when the number of p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com