Method for quickly and accurately detecting copy number variation of CYP2D6 gene
A gene and detection object technology, applied in the field of determining the copy number change of CYP2D6 gene from small groups or individuals, can solve the problems of low sensitivity, high reagent cost, long detection time, etc., achieve simple and fast operation, improve accuracy, Effects that are easy to scale
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
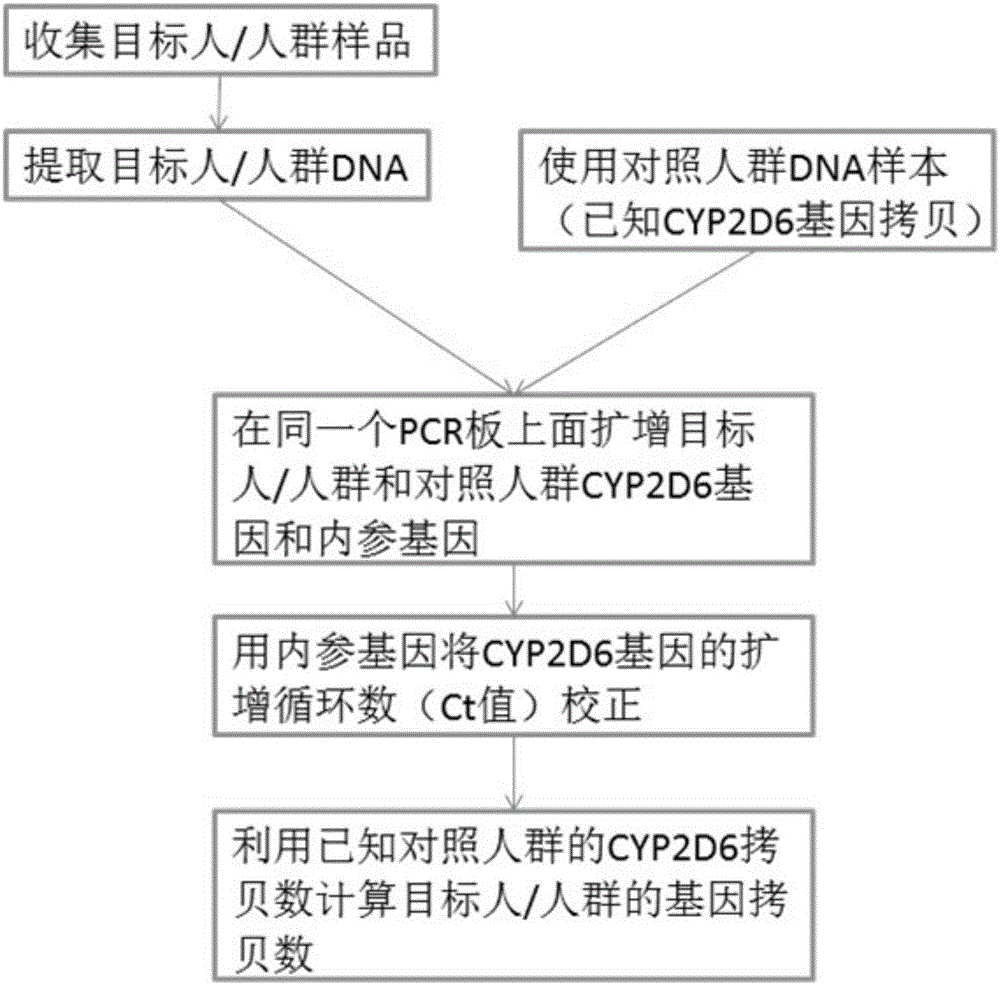
Image
Examples
Embodiment 1
[0038] Example 1 Preparation of primers for detecting CYP2D6 gene polymorphisms:
[0039] Including PCR amplification primers and fluorescent probes for 3 segments of CYP2D6 gene. The corresponding probe of CYP2D6 gene was labeled with FAM at the 5' end.
[0040] The sequences of the three sets of primers and probes for the specific CYP2D6 gene are as follows:
[0041]
[0042]
Embodiment 2
[0043] Example 2 Preparation of RPP30 gene primers and probes:
[0044] In order to study the copy number variation of CYP2D6 gene, the human genome single-copy gene RPP30 (ribonuclease P / MRP 30kDa subunit) was selected as the reference sequence. The corresponding probe of RPP30 gene was labeled with HEX at the 5' end.
[0045] RPP30 gene primers and probes are:
[0046]
Embodiment 3
[0047] Embodiment 3 fluorescence quantitative PCR reaction:
[0048]The copy number variation of CYP2D6 gene is detected by single-tube double quantitative PCR method. The specific reaction system is 10 μL, including 8ng gene DNA, 5 μL 2x TaqMan mix, RPP30 gene primers and probes, and a set of CYP2D6 gene primers and probes (The final concentration of both primers and probes is 0.2 μM), and then add water to 10 μL. For each sample, 4 replicates were performed. The PCR amplification conditions were hot start at 95°C for 10 min, denaturation at 95°C for 15 s, annealing and amplification at 60°C for 1 min, a total of 40 cycles.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com