Molecular marker related to weaning weight and anti-diarrheal diseases of piglets and application of molecular marker in pig breeding
A piglet weaning and molecular marker technology, applied in the field of molecular genetics, to achieve the effect of improving weaning weight and resistance to diarrheal disease, increasing weaning weight of piglets, and improving resistance to diarrheal disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
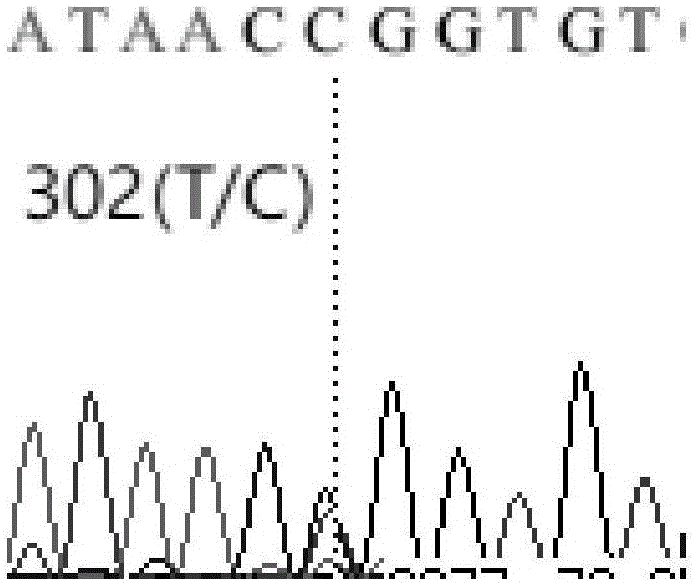
[0049] Example 1: Obtaining of molecular markers related to piglet resistance to diarrheal disease and weaning weight:
[0050] 1) Primer design and partial DNA sequence amplification of porcine IL6 gene
[0051] Download porcine IL6 gene mRNA sequence (GeneBank accession number: NM_214399.1) and genome sequence (GeneBank accession number: NC_010451.3 REGION: complement (101081042..101085451) GPC_000000591, shown in SEQ ID NO.1) from NCBI. The two sequences were compared, and the downloaded sequence was (NC_010451.3REGION:complement(101081042..101085451)GPC_000000591, shown in SEQ ID NO.1) model sequence, and Primer5 software was used to design primer IL6-3 for amplifying IL6 Part of the gene Inron2, Intron3 and the complete Exon3 region, primer IL6-5 was designed to amplify part of the Intron4 and part of the Exon5 region of the IL6 gene, all primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd., and the primer sequences are listed in Table 1 below Show:
...
Embodiment 2
[0059] Embodiment 2: the detection method of HpaII site, its steps are as follows:
[0060] 1) Primer sequence
[0061] The primers used for PCR amplification are IL6-3 primers, and the primer sequences are as follows
[0062] IL6-3F: 5'TGCGAATGGGTTCTGACT 3';
[0063] IL6-3R: 5'AATCGCTCTTCCCTCTGG 3'
[0064] 2) PCR reaction conditions
[0065] Use IL6-3 primers to amplify in porcine genomic DNA. The PCR reaction system is 25 μL, including 1 μL of template DNA, 0.5 nmol / μL of upstream and downstream primers, 12.5 μL of PCR mix (Bao Biology), and add deionized water to the total volume. 25 μL. The PCR program was as follows: pre-denaturation at 94°C for 4 min; then denaturation at 94°C for 35 s, annealing at 53°C for 35 s, and extension at 72°C for 40 s, a total of 35 cycles; finally, extension at 72°C for 8 min.
[0066] 3) HpaII PCR-RFLP detection
[0067] The total PCR product digestion reaction system is 10 μl, including 8.5 μl of PCR product, 0.5 μl (10 U / μl) of HpaII...
Embodiment 3
[0068] Embodiment 3: the detection method of DraI site, its steps are as follows:
[0069] 1) Primer sequence
[0070] The primers used for PCR amplification are IL6-5 primers, and the primer sequences are as follows
[0071] IL6-5F:5'CAAATGTGAACCTCCTCC 3'
[0072] IL6-5R:5'AACAACCTGGCTCTGAAA 3'
[0073] 2) PCR reaction
[0074] Use IL6-5 primers to amplify in porcine genomic DNA. The PCR reaction system is 25 μL, including 1 μL of template DNA, 0.5 nmol / μL of upstream and downstream primers, 12.5 μL of PCR mix (Bao Biology), and add deionized water to the total volume. 25 μL. The PCR program was as follows: pre-denaturation at 94°C for 4 min; then denaturation at 94°C for 35 s, annealing at 50°C for 35 s, and extension at 72°C for 40 s, a total of 35 cycles; finally, extension at 72°C for 8 min.
[0075] 3) DraI PCR-RFLP detection
[0076] The total PCR product digestion reaction system is 10 μl, including 8.5 μl of PCR product, 0.5 μl (10 U / μl) DraI endonuclease, and 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com