A miR-126 full-length gene knockout kit based on CRISPR-Cas9 technology and its application
A mir-126 and full-length gene technology, applied in the field of miR-126 gene knockout kits, can solve the problems of adverse effects on research, inaccurate sequence insertion and deletion, and the repair process is not under human control, etc., to overcome time-consuming The effects of cumbersomeness, reduced reagent cost consumption, simple construction steps and experimental operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1. Vector construction
[0072] (1) miR-126 target design
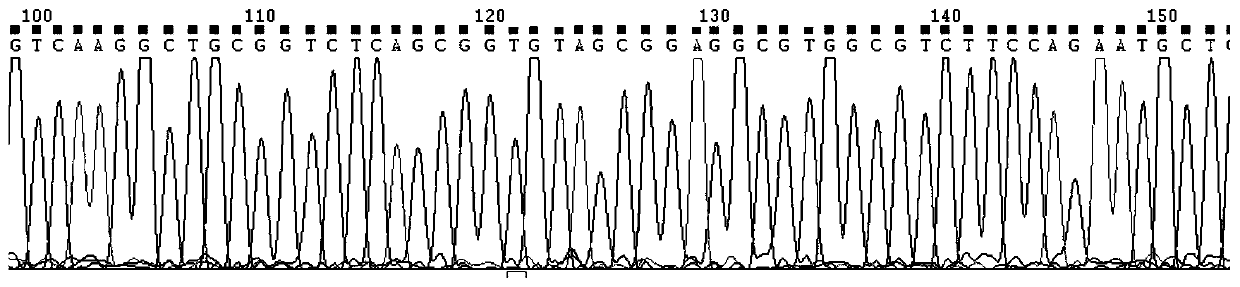
[0073] For the miR-126 gene (gene name miR-126, gene ID number: 406913, see http: / / www.ncbi.nlm.nih.gov / gene / 406913 for gene details), download miR-126 from this website Genome sequence:
[0074] 5'-CGCTGGCGACGGGACATTATTACTTTTGGTACGCGCTGTGACACTTCAAACTCGTACCGTGAGTAATAATGCGCCGTCCACGGCA-3', as shown in SEQ ID NO.1.
[0075] Use the online software Feng Zhang lab's Target Finder (http: / / crispr.mit.edu / ) to design sgRNA, input the miR-126 genome sequence and its upstream and downstream 50bp sequences, set and retrieve several sgRNA sequences, and analyze the sgRNA in The location on the gene sequence and the off-target (off-target) information of the sgRNA, from which the optimal 2 upstream target sequences and 3 downstream target sequences are respectively selected, as follows:
[0076] upstream:
[0077] sgRNA-1: TAATGTCCCGTCGCCAGCGG, as shown in SEQ ID NO.2;
[0078] sgRNA-2: GCCACGCCTCCGCTGGCGAC, as...
Embodiment 2
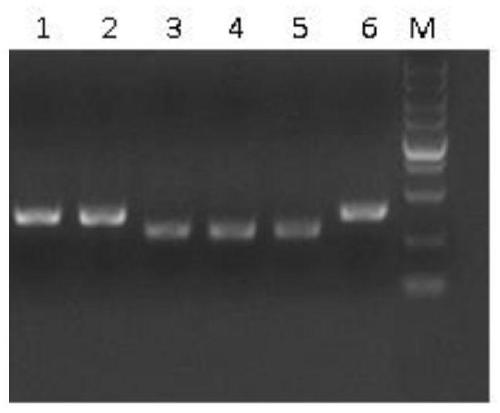
[0111] Example 2. Plasmid transfection and identification of miR-126 knockout activity in 293T cells and construction of 293T knockout miR-126 cell model
[0112] (1) Plasmid extraction
[0113] The Escherichia coli DH5α of the pSpCas9(BB)-2A-Puro plasmid respectively comprising sgRNA-1, sgRNA-2, sgRNA-3, sgRNA-4, and sgRNA-5 fragments verified by sequencing in Example 1 were each inoculated in 5 mL In LB liquid medium, culture overnight at 37°C with shaking. Plasmid extraction was performed on these bacterial liquids using an endotoxin-free mini-plasmid extraction kit (the kit was produced by Omega Company, the article number is D6948-02). The extracted plasmid is then precipitated with absolute ethanol in an ultra-clean bench, and then dissolved in sterile water for aseptic treatment of the plasmid.
[0114] (2) 293T cells were transfected with a pair of pSpCas9(BB)-2A-Puro-sgRNA plasmids containing sgRNA
[0115] 1) 293T cell culture
[0116] 293T cells were divided int...
Embodiment 3
[0152] Example 3 Construction of miR-126 knockout lung cancer cell model
[0153] 1) Transfection of lung cancer cells A549 cells
[0154] Co-transfect lung cancer cell A549 with pSpCas9(BB)-2A-Puro-sgRNA1 and pSpCas9(BB)-2A-Puro-sgRNA3 using Lipofectamine 2000. After 24 hours of transfection, use the complete medium containing 2 μg / mL puromycin to carry out drug treatment. After three days of drug sieving, the cells were collected.
[0155] 2) Knockout activity verification
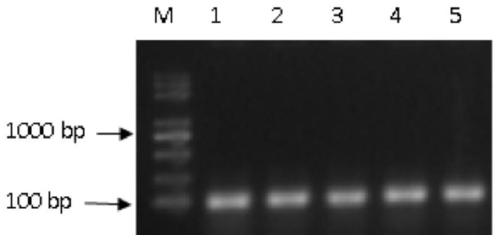
[0156] Collect the A549 cells after the drug screening and the A549 cells without transfection, use the ultra-micro sample genotype identification kit (Nanjing Yaoshunyu Biotechnology Co., Ltd., KC-101) for sample processing, and use as shown in Table 10 PCR primers for PCR amplification identification.
[0157] Table 10
[0158] *Primer sequence (5'--3') *Primer name GAGGGAGGATAGGTGGGTTC, as shown in SEQ ID NO.19 mir126-test-Fw AGGCAGAGCCAGAAGACTCA, as shown in SEQ ID NO.20 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com