Novel phosphatidase B and application thereof
A technology of phospholipase and application, which is applied in the field of genetic engineering of enzymes, and can solve problems such as difficult to obtain satisfactory results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
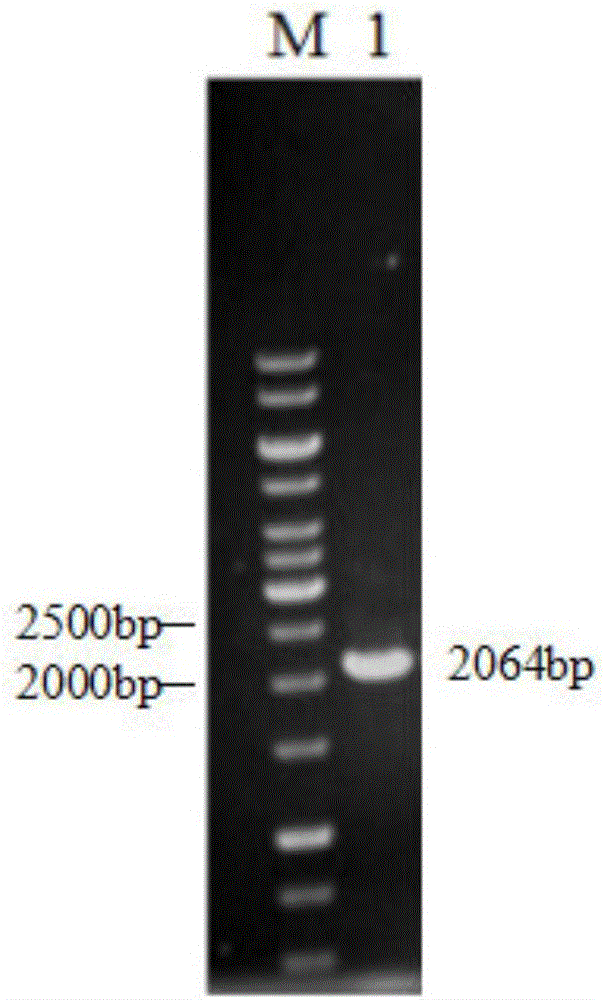
[0052] Embodiment 1: Obtaining of wild-type phospholipase B gene
[0053] 1. The wild-type phospholipase B gene plb is from Saccharomyces cerevisiae TCCC31028, and its genomic DNA is extracted, wherein the extraction steps of Saccharomyces cerevisiae genomic DNA are as follows:
[0054] (1) Pick a ring of bacteria from the culture plate and inoculate in 50mL of appropriate medium, culture at 30°C and 250r / min for 16-18h.
[0055] (2) Afterwards, take 1 mL of the culture solution in a 1.5 mL EP tube, centrifuge at 12,000 r / min for 2 min, pour off the supernatant, resuspend with 200 μL of lysate, add quartz sand, and shake for 20-30 min.
[0056] (3) Add 500uL of lysate, centrifuge at 12000r / min for 5min, and take the supernatant.
[0057] (4) Add an equal volume of Tris to balance phenol:chloroform=1:1, mix well, centrifuge at 12000rpm for 5min, and transfer the supernatant to another EP tube.
[0058] (5) Repeat the extraction twice until no protein layer appears, and finall...
Embodiment 2
[0075] Embodiment 2: the acquisition of phospholipase B mutant gene plbm
[0076] 1. Random mutation based on error-prone PCR technology
[0077] In the error-prone PCR reaction system, P1 and P2 were used as upstream and downstream primers, and plb was used as a template to perform error-prone PCR to obtain plbm.
[0078] The reaction conditions for its amplification are:
[0079]
[0080]
[0081] The amplification conditions were: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 30 s, annealing at 70°C for 40 s, extension at 72°C for 2 min, and 30 cycles of reaction; extension at 72°C for 10 min.
[0082] 2. The phospholipase B error-prone PCR product is cloned into the expression vector pET28a, transformed into Escherichia coli BL21, inoculated in a 96-well cell culture plate containing 200 μL of LB liquid medium (containing 30 μg / mL of Kan) in each well, at 37 Cultivate on a shaker at 200r / min under the condition of ℃, when the OD600 reaches 0.6, add I...
Embodiment 3
[0101] Example 3: Construction of Pichia pastoris free expression recombinant bacteria
[0102] 1. Construction of wild-type phospholipase B and phospholipase B mutant expression vectors pPIC9K-plb and pPIC9K-plbm
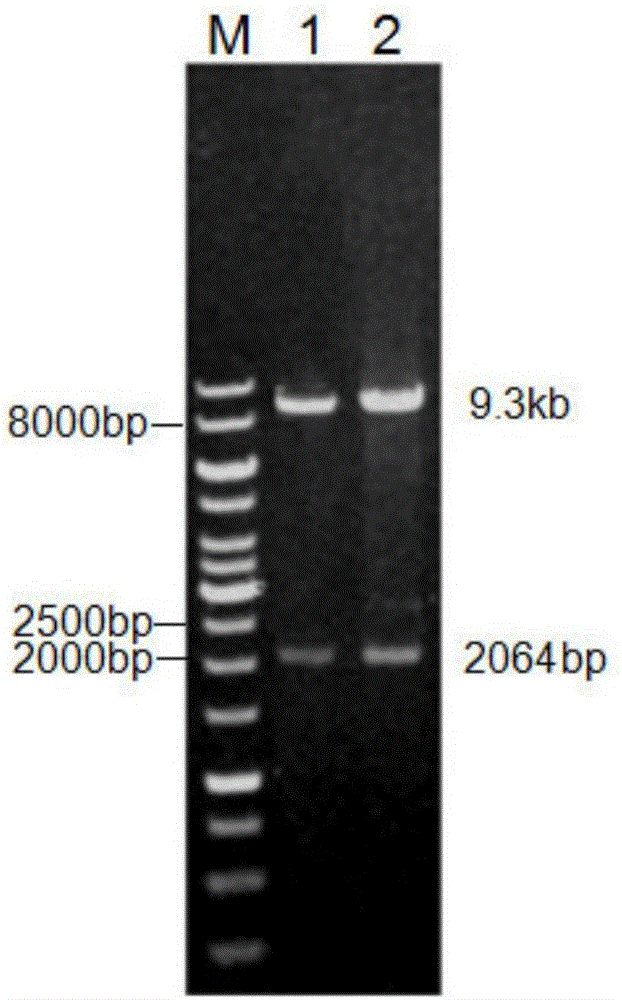
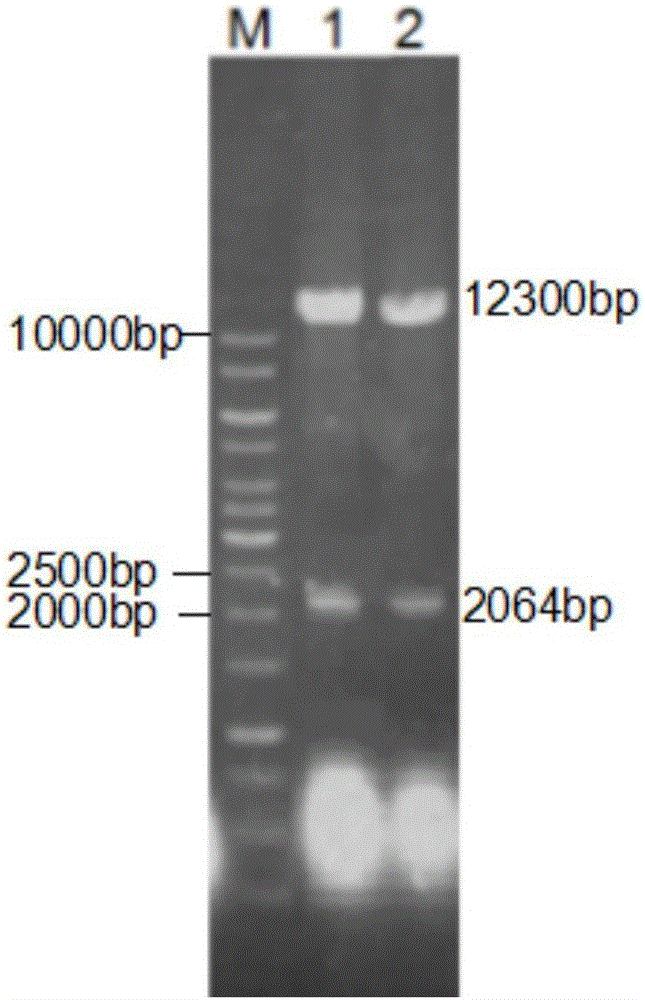
[0103] plb and plbm were respectively digested with Pichia pastoris expression vector pPIC 9K by EcoRI and NotI, then ligated, transformed into Escherichia coli JM109 competent cells, and Amp was selected r The positive transformant, the plasmid was extracted after colony culture, and the enzyme digestion verification was successful (see figure 2 ) to obtain recombinant expression vectors pPIC9K-plb and pPIC9K-plbm.
[0104] 2. Construction of wild-type phospholipase B and phospholipase B mutant expression recombinant strains
[0105] (1) Linearization of plasmid DNA
[0106] Before transforming Pichia pastoris, the recombinant expression plasmids pPIC9K-plb and pPIC9K-plbm were linearized with SalI restriction endonuclease.
[0107] (2) Electroporation of lin...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extend | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com