Cotton somatic cell chromosome Oligo-FISH (oligonucleotide-fluorescence in situ hybridization) method
A chromosome and somatic cell technology, applied in the fields of bioinformatics and molecular cytogenetics, can solve problems such as difficulty in guaranteeing efficiency and stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The present invention will be further described below in conjunction with accompanying drawing:
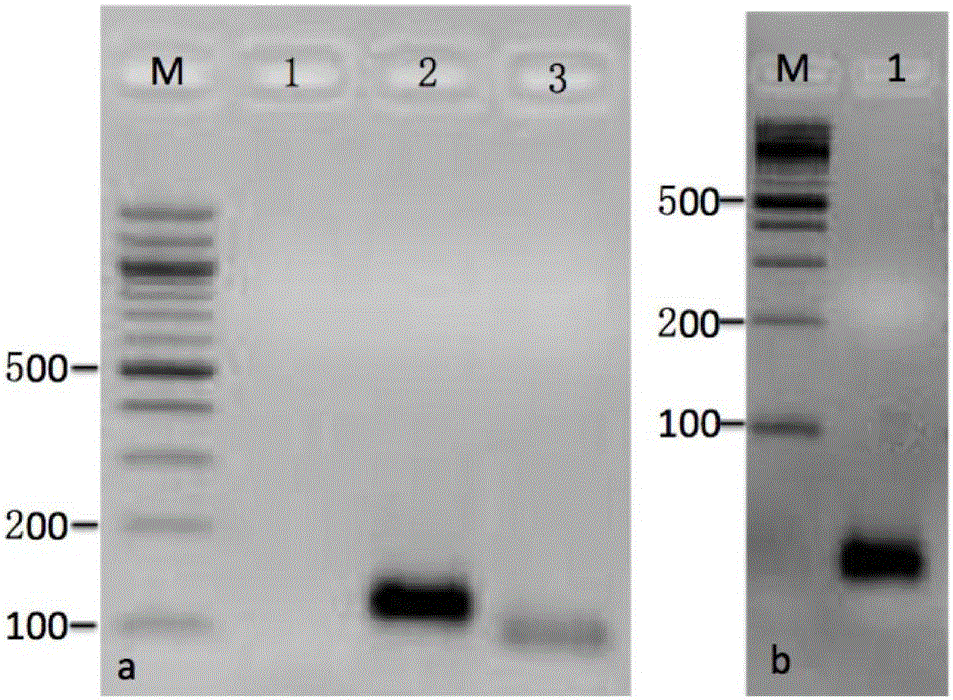
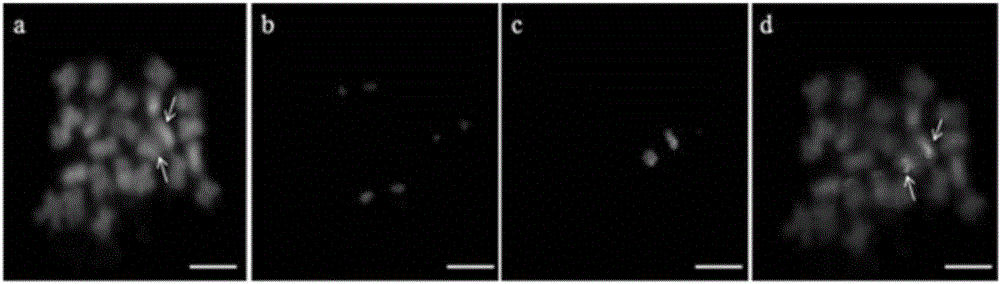
[0028] Such as figure 1 Shown: the genome of the oligos designed in the present invention is the diploid D genome wild species Raymond's cotton G. raimondii (D5) genome (Paterson et al.2012); the cotton material used for in situ hybridization is the diploid D genome Ray G. raimondii (D5) was planted in the Hainan wild cotton plantation of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences in Sanya, Hainan, and had a backup in the greenhouse of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences in Anyang, Henan; the specific methods include Raymond Cotton No. Design of oligonucleotide mixing pool for chromosome 1 and labeling of oligonucleotide probes;
[0029] The oligonucleotide mixed pool design of the No. 1 chromosome of Raymond's cotton comprises the following steps:
[0030] (1) Use the RepeatMasker system to remo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com