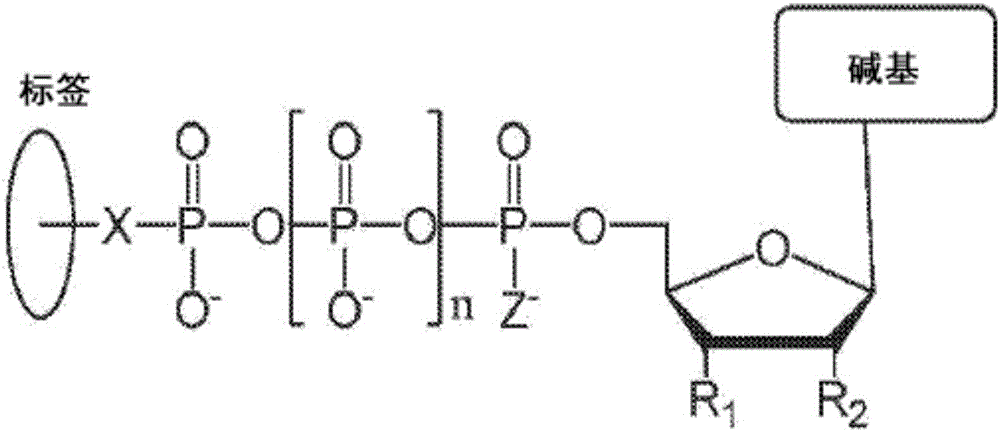
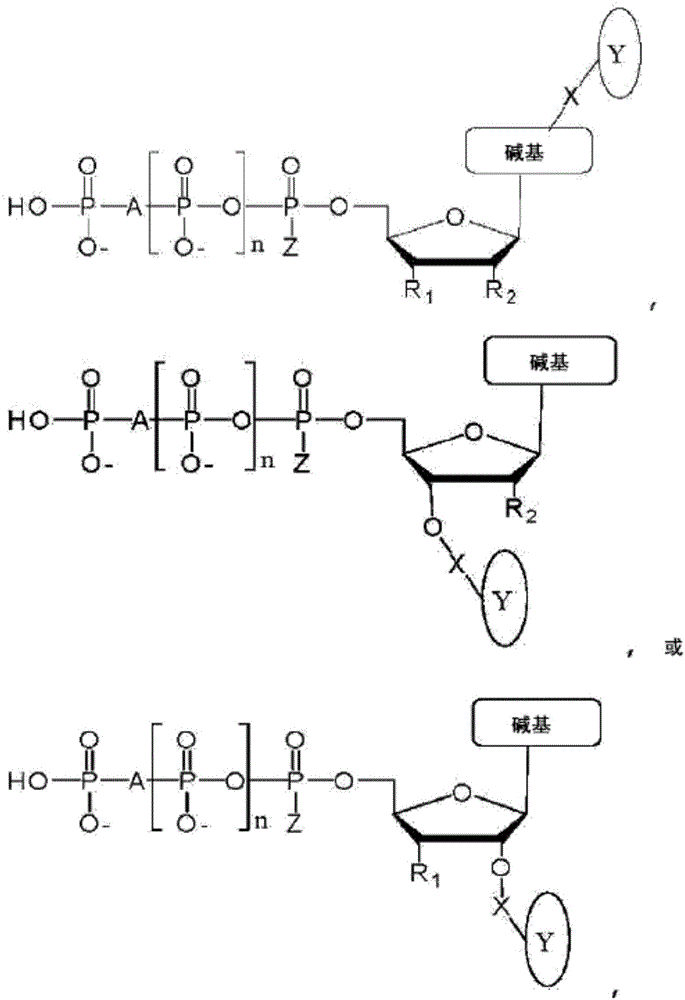
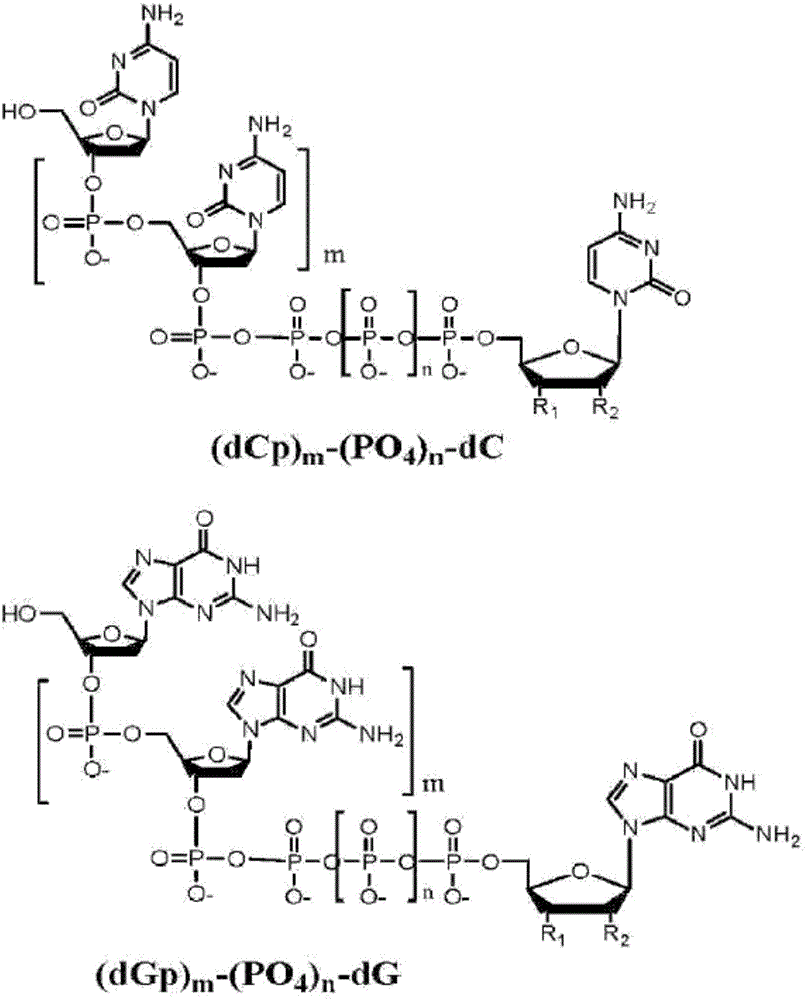
Chemical methods for producing tagged nucleotides
A nucleotide and tagged technology, which can be used in biochemical equipment and methods, organic chemistry, microbial measurement/testing, etc., and can solve the problem of insufficient signal difference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0268] Example 1 - Synthesis of Coumarin-PEG-dG4P Tagged Nucleotides
[0269] In this example, nucleotides were purified by reverse phase HPLC using a 150 x 4.6 mm column (Supelco), mobile phase: A, 8.6 mM Et 3 N / 100 mM 1,1,1,3,3,3-hexafluoro-2-propanol in water (pH 8.1); B, methanol. Elution was performed as follows: 100% A isocratic for 10 minutes, followed by a linear gradient of 0-50% B for 20 minutes, then 50% B for 30 minutes.
[0270] Such as Figure 16 As shown in the scheme, Coumarin-PEG n The synthesis of -dG4P includes three synthetic operations: A, B and C.
[0271] A. 2'-Deoxyguanosine-5'-tetraphosphate (dG4P) and dG4P-NH 2 Synthesis: First, 2'-dG4P was synthesized from 2'-dGTP. 300 μmol of 2'-dGTP (triethylammonium salt) was converted to the tributylammonium salt by using a solution of 1.5 mmol (5 equivalents) of tributylamine in dry pyridine (5 ml). The resulting solution was concentrated to dryness and co-evaporated with 5 ml anhydrous DMF (x2). ...
Embodiment 2
[0281] Example 2 - Characterization of released tags by MALDI-TOF MS
[0282] After HPLC purification, the expected coumarin-PEG-NH was confirmed by MALDI-TOF-MS analysis 2 molecular( Figure 17 ). MALDI-TOF-MS results showed that the coumarin-PEG-NH produced by acid hydrolysis 2 The tag is identical to the released tag produced during the polymerase reaction following alkaline phosphatase treatment.
[0283] refer to Figure 17 , as shown by MALDI-TOF-MS analysis, by generating coumarin-PEG16-NH 2 Acid hydrolysis of coumarin-PEG16-dG4P yields coumarin-PEG20-NH 2 Acid hydrolysis of coumarin-PEG20-dG4P yields coumarin-PEG24-NH 2 Acid hydrolysis of coumarin-PEG24-dG4P and generation of coumarin-PEG36-NH 2 Coumarin-PEG-NH produced by acid hydrolysis of coumarin-PEG36-dG4P 2 Tag, identical to the corresponding release tag generated in the polymerase extension reaction after treatment with alkaline phosphatase. A composite image of four individually acquired MS spectra i...
Embodiment 3
[0284] Example 3—Detection of oligonucleotide labels
[0285] Nanopore array device (see for example Figure 12 ) for detecting 4 different current levels for 4 different labels. Such as Figure 18 As shown, each tag can be distinguished from any of the other three tags (ie, the histogram shows four distinct peaks labeled with the corresponding tag in the graph). Each tag is a "T" oligonucleotide homopolymer approximately 30 bases in length, biotinylated at the 3' end, with two regions in the potentially modified strand. In each 30-base long molecule, the modified regions were base positions 11, 12 and 13 and positions 17, 18 and 19 from the 3' end. As used herein, "x" is an abasic site (abasic), and "T" is thymine. The four tags are:
[0286] (a) "pseudo-label-XXX_XXX" has the sequence: streptavidin-biotin-10T-xxx-3T-xxx-11T (SEQID NO.1),
[0287] (b) "pseudo-tag-TTT_XXX" has the sequence: streptavidin-biotin-10T-TTT-3T-xxx-11T (SEQID NO.2),
[0288] (c) The "30T" ta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com