Method for obtaining stable cell populations for expressing target protein in CHO cells through piggyBac transposons
A target protein and transposon technology, applied in the field of biopharmaceuticals, can solve the problems of difficult to adapt to the construction time, slow recovery of cell groups, low production efficiency, etc., so as to save the cost of liquid nitrogen storage and management and cell expression Stable, easy-to-operate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
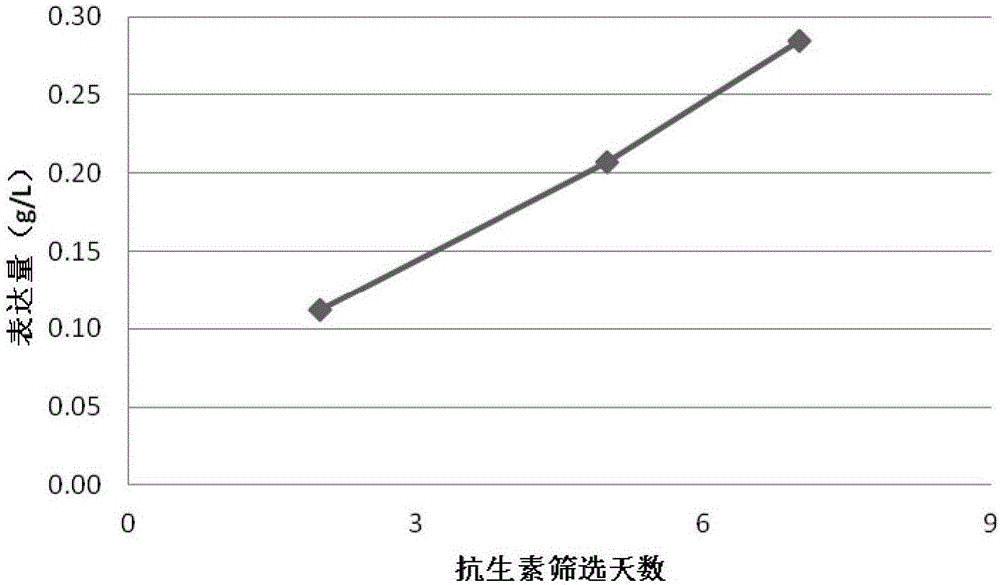
[0030] In this example, piggyBac transposons were used to construct different stable cell populations. After 2 days, 5 days or 7 days of antibiotic selection, batch feeding experiments were performed to express proteins, so as to obtain a suitable antibiotic selection time. Specifically, the following steps are included:
[0031] Step 1, the gene of the target protein and the antibiotic gene are cloned between a terminal inverted repeat sequence recognized by a pair of piggyBac transposase. For target proteins containing multiple expression units, such as antibodies or Fc fusion proteins, different expression units can be cloned into vectors containing different antibiotic genes.
[0032] Step 2, use the endotoxin-free plasmid extraction kit to extract the plasmids containing transposase and target protein respectively. All plasmids do not need to be digested and linearized.
[0033] In step 3, the host cells are CHO cells that have been acclimated to suspension. The expect...
Embodiment 2
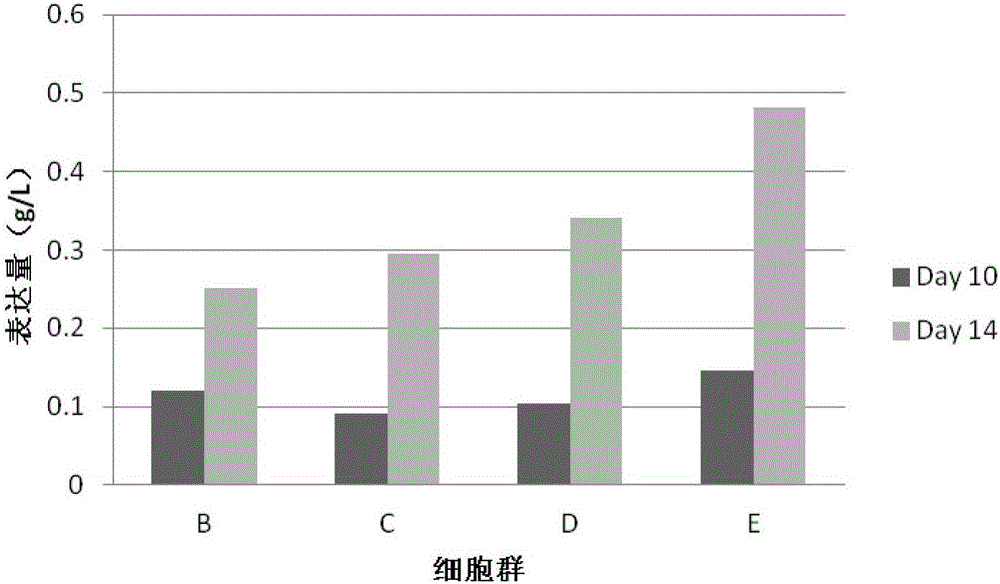
[0042] In this example, piggyBac transposons were used to construct four different stable cell populations of B, C, D, and E encoding human IgG1 antibodies. After being screened with antibiotics for one week, a batch fed-feed experiment was performed to express the protein, and the number of feeding days was recorded as 10 days and 14-day protein expression data. Specifically, the following steps are included:
[0043] Step 1, the gene of the target protein and the antibiotic gene are cloned between a terminal inverted repeat sequence recognized by a pair of piggyBac transposase. For target proteins containing multiple expression units, such as antibodies or Fc fusion proteins, different expression units can be cloned into vectors containing different antibiotic genes.
[0044] Step 2, use the endotoxin-free plasmid extraction kit to extract the plasmids containing transposase and target protein respectively. All plasmids do not need to be digested and linearized.
[0045] ...
Embodiment 3
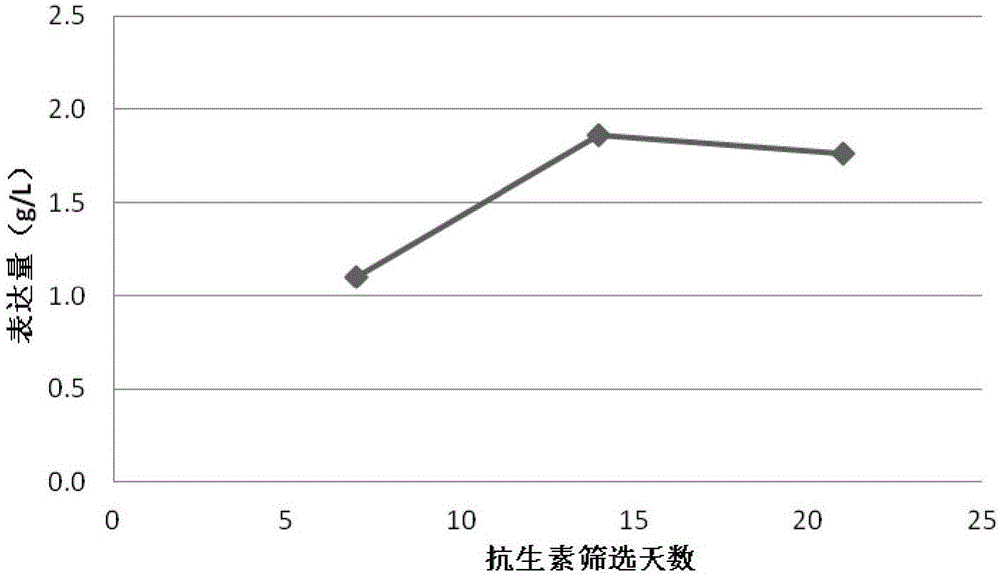
[0054] This example estimates the applicable production volumes of this method by evaluating the stability of expression in stable cell populations constructed with piggyBac transposons. Specifically, the following steps are included:
[0055] Step 1, the gene of the target protein and the antibiotic gene are cloned between a terminal inverted repeat sequence recognized by a pair of piggyBac transposase. For target proteins containing multiple expression units, such as antibodies or Fc fusion proteins, different expression units can be cloned into vectors containing different antibiotic genes.
[0056] Step 2, use the endotoxin-free plasmid extraction kit to extract the plasmids containing transposase and target protein respectively. All plasmids do not need to be digested and linearized.
[0057] In step 3, the host cells are CHO cells that have been acclimated to suspension. On the day of transfection, the host cell density is expected to be between 1 and 2E6 / ml.
[0058...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com