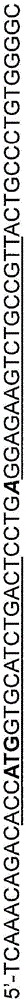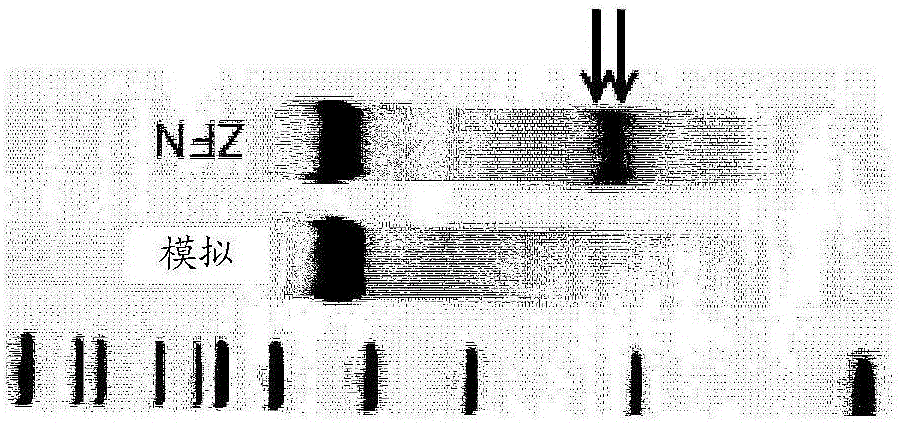Methods and compositions for nuclease-mediated genome engineering and correction in hematopoietic stem cells
A hematopoietic stem cell and genome technology, applied in the field of genome engineering of hematopoietic stem cells, can solve the problems of life expectancy patients' quality of life, unknown long-term side effects, unwanted side effects, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0180] Example 1: Design, Construction and General Characterization of Zinc Finger Nucleases (ZFNs)
[0181] Zinc finger proteins were designed and incorporated into plasmid, AAV or adenoviral vectors essentially as Urnov et al. (2005) Nature 435(7042):646-651, Perez et al (2008) Nature Biotechnology 26(7) : 808-816, and as described in U.S. Patent No. 6,534,261 and tested for binding. See commonly owned US Patent No. 7,888,121 and US Patent Publication Nos. 20130137104 and 20130122591 for ZFNs and TALENs specific for the human beta globin locus.
Embodiment 2
[0182] Example 2: Activity of globin-specific ZFNs
[0183] Pairs of ZFNs targeting the human globin locus were used to test the ability of these ZFNs to induce DSBs at specific target sites. The amino acid sequences of the recognition helical regions of each finger of the indicated ZFNs are shown in Table 1 below. Target sites (DNA target sites are indicated in capital letters; non-contact nucleotides are indicated in lower case letters) are shown in Table 2.
[0184] Table 1: Human β-globin-specific zinc finger protein recognition helix design
[0185]
[0186]
[0187] Table 2: Target sites of human β-globin-specific zinc fingers
[0188]
[0189] Human CD34+ cells were obtained and processed as follows. All cord blood (CB) specimens were obtained according to UC-approved guidelines and were considered anonymous medical waste exempt from IRB review. Process cells within 48 hours of collection. Bone marrow (BM) aspirates from volunteer donors with SCD were obta...
Embodiment 4
[0201] Example 4: ZFN-driven correction at sickle cell bases
[0202] Following successful cleavage of the targeted β-globin locus, we sought to determine whether it was possible to correct sickle bases at this locus using a cognate donor template. To this end, two types of gene correction templates were simultaneously designed and tested: short DNA oligonucleotides and IDLV. The 1.1 kb human β-globin gene fragment donor template cloned into IDLV was designed to include correction changes for sickle mutations as well as silent restriction fragment length polymorphisms (RFLPs) to generate replacements for homologous recombination The analyzed HhaI restriction site ( Figure 1C ). Donor templates are delivered via IDLV (see Figure 2D ), enabling efficient transduction of CD34+ HSPCs with minimal cytotoxicity, resulting in transient generation of high template copy numbers with minimal genomic integration.
[0203] Levels of gene modification in CD34+ cells treated with ZFN ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com