Multiple PCR (Polymerase Chain Reaction) primer groups and detection method for synchronously detecting four pathogenic bacteria of siluriformes fish
A detection method and technology for pathogenic bacteria, which are applied in biochemical equipment and methods, recombinant DNA technology, and microbial determination/inspection, etc. Rapid identification, good sensitivity, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The extraction of embodiment 1 DNA
[0047] Genomic DNA was extracted using a DNA kit as a template
Embodiment 2
[0048] Embodiment 2 Establishment of multiplex PCR amplification method
[0049] Firstly, through single gene-specific PCR, the reaction system and reaction conditions for each pair of primer sets to amplify the corresponding gene fragments were preliminarily explored. The genomic DNAs of the four pathogenic bacteria were extracted separately as the amplification templates of the respective primer sets. Through multiple experiments, it was found that each pair of primer sets can well amplify the corresponding gene fragments under the following reaction systems and conditions.
[0050] 1. Establishment of single-gene PCR reaction system and reaction conditions
[0051] 1), the system and conditions of the single-gene-specific PCR reaction of Edwardsiella catfish
[0052] PCR reaction system:
[0053]
[0054] PCR reaction conditions: pre-denaturation at 98°C for 5min, denaturation at 98°C for 30s, annealing at 57-60°C for 30s, extension at 72°C for 1min, 30 cycles, and fin...
Embodiment 3
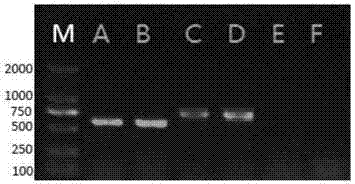
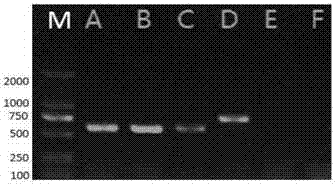
[0080] The electrophoretic detection of embodiment 3 PCR amplification product
[0081] Use conventional electrophoresis buffer TAE buffer (0.04mol / L Tris-acetic acid, 0.001mol / L EDTA) to configure 1.2% agarose gel. After sample application, set the voltage to 100-140 volts and perform electrophoresis for 20-40 minutes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com