RT-PCR (Reverse Transcription-Polymerase Chain Reaction) method for detecting porcine reproductive and respiratory syndrome virus (PRRSV) classical strains, high-pathogenicity variant strains and NADC-30 strains
A porcine blue ear virus, RT-PCR technology, applied in biochemical equipment and methods, DNA / RNA fragments, recombinant DNA technology, etc., can solve the problems of long detection time and complicated operation, and achieve high sensitivity and few operation steps , the effect of saving time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Primer Design
[0024] According to the three subtype sequences of the PRRSV gene registered in NCBI (classic strain of porcine PRRS virus (EF536003), highly pathogenic variant strain (EF112445) and NADC-30 strain (JN654459)), the software design was applied Primer5.0 For the Nsp2 gene of PRRSV, specific primers that can distinguish classic strains of porcine PRRSV, highly pathogenic variant strains, and NADC-30 strains were used. All primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd.
[0025] Upstream primer (Nsp2F primer): GACACCTCCTTTGATTGG, SEQ ID NO: 1;
[0026] Downstream primer (Nsp2R primer): GAGAGGAIGCAGACAAATC (where I represents T or C or A or G), SEQ ID NO: 2;
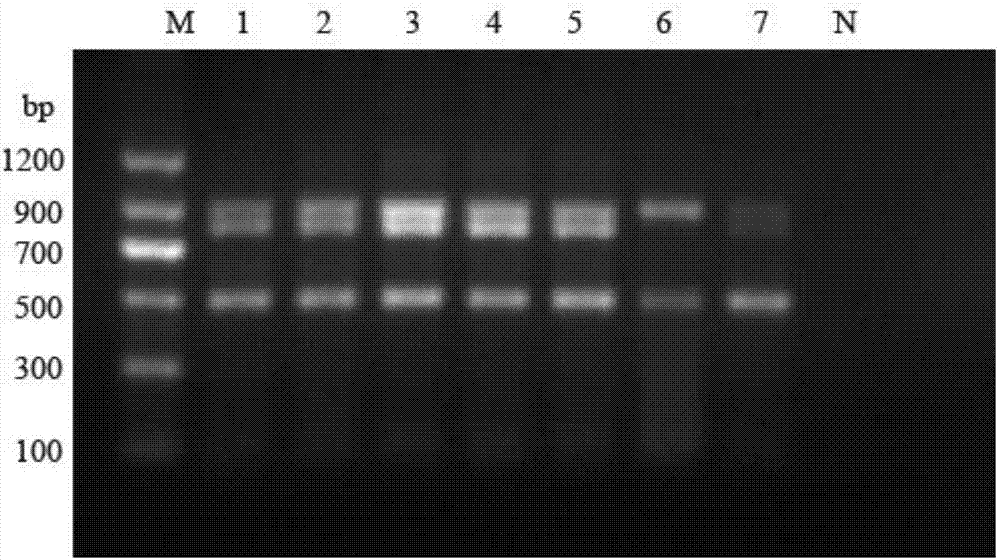
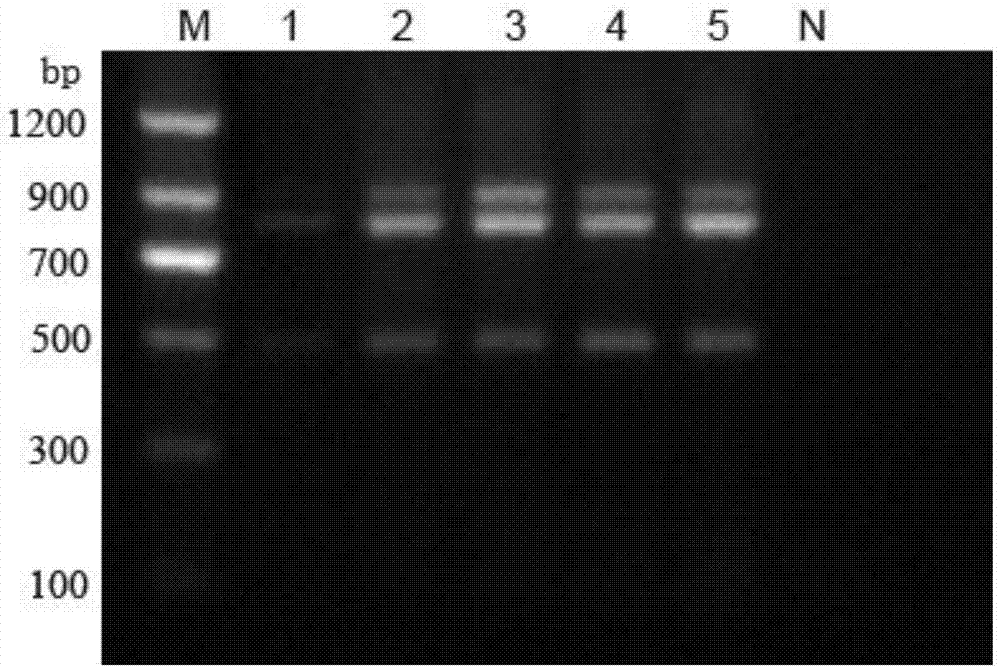
[0027] Using above-mentioned primer RT-PCR amplification, the three subtypes of PRRSV can be quickly identified and distinguished by the size of the agarose gel electrophoresis band, the specificity and sensitivity are high, and the operation is simple.
Embodiment 2
[0028] Embodiment 2 detects the RT-PCR method of porcine PRRS classic strain, highly pathogenic variant strain and NADC-30 strain
[0029] The method includes the following steps:
[0030] (1) RNA extraction of samples to be tested:
[0031] The blood samples to be tested are centrifuged at 8000g for 5 minutes, and the upper serum is taken for each use; the tissue and organ samples need to be mixed with sterilized double distilled water at a mass volume ratio of 1:5, centrifuged at 8000g for 5 minutes after grinding, and the supernatant is taken for later use.
[0032] (2) The extracted RNA was added to a reverse transcription kit purchased from Bao Bio Engineering (Dalian) Co., Ltd. for reverse transcription to obtain a cDNA template:
[0033] The reverse transcription step is: mix 4 μL of RNA template with No. 1 5×PrimeScript Buffer 4 μL, No. 2 PrimeScriptRT Enzyme Mix 1 μL, No. 4 Random 6mers 2 μL and No. 5 RNase Free dH 2 O 9 μL was mixed and put into a PCR machine for r...
Embodiment 3
[0037] The optimization of embodiment 3 annealing temperature and primer concentration and the mensuration of sensitivity
[0038] (1) Cloning transformation and recombinant positive plasmid: RT-PCR amplifies the target gene, and conducts electrophoresis observation and analysis. The positive samples screened by electrophoresis were purified by the PCR purification kit purchased from OMGA Company, and the purified PCR products were connected to the TMD-19T carrier, put in the metal bath for 12 hours, and then taken out. The obtained ligation product was added to competent cells (Escherichia coli DH5α strain), and then the liquid was added to 1 mL of LB liquid, placed in a 37°C shaker box, and then centrifuged at 5000r / min for 4min, 800 μL of the supernatant was discarded, and the remaining 200 μL was blown and mixed. Take the bacterial liquid and drop it on the medium of LB+AMP, and spread the bacterial liquid evenly on the medium with a spreader stick, and put it in a 37°C ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com