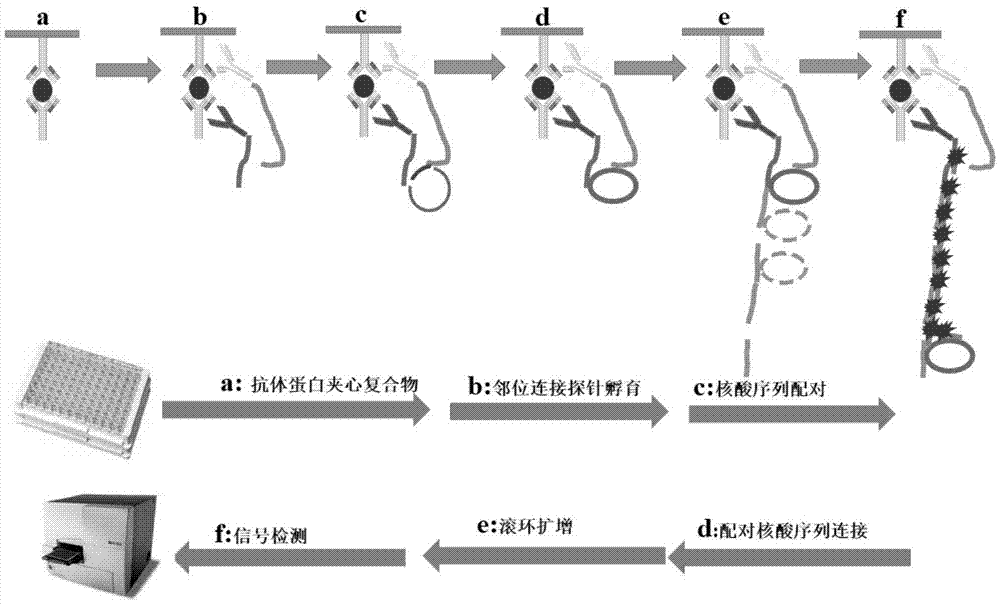
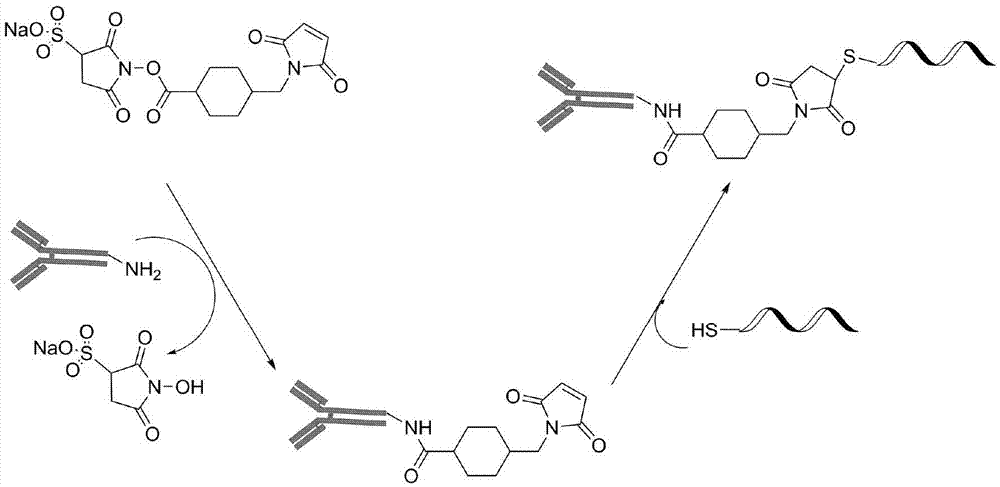
Method for quantitatively detecting low-abundance protein and post-modification proteinthereof
A low-abundance protein, quantitative detection technology, applied in the direction of measuring devices, biological testing, material inspection products, etc., to achieve rapid and sensitive specific detection, reduce background interference, and improve specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
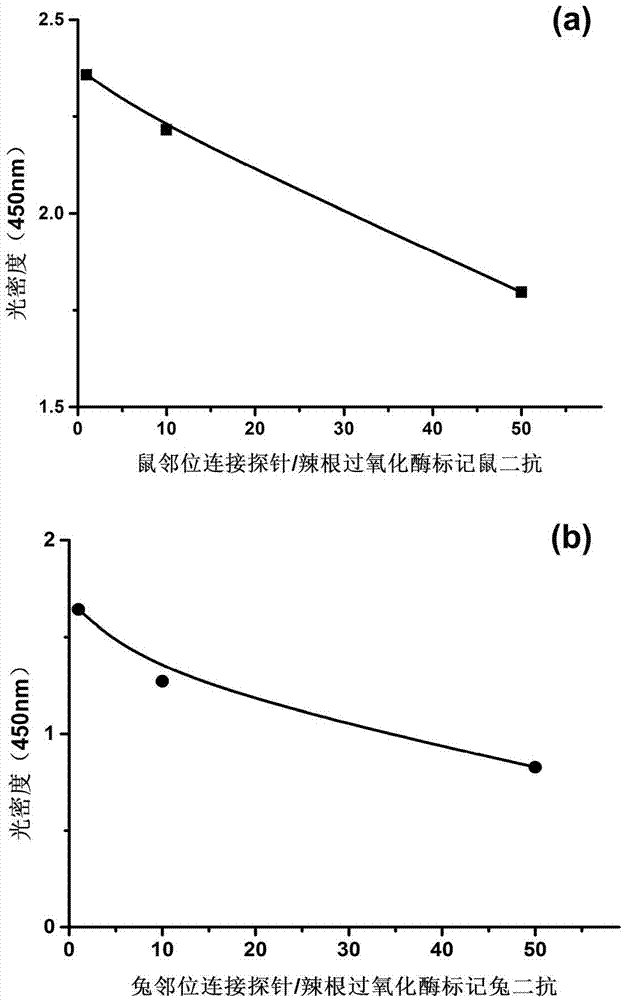
[0038] The investigation of the activity of embodiment 1 synthetic proximity ligation probe
[0039] Proximity ligation probes were synthesized as figure 2 Shown: Take 20ug rabbit secondary antibody and 20ug mouse secondary antibody dissolved in 10μl phosphate buffer (PH7.4), respectively add 0.5μl 8mM sulfo-SMCC, react at room temperature for 2 hours; Ultrafilter the filter tube four times to remove excess sulfo-SMCC; add 4 μl of 100 uM thiol-modified primer nucleic acid sequence to the ultrafiltered mouse secondary antibody, and add 4 μl of 100 μM thiol-modified non-primer nucleic acid sequence to the ultrafiltered rabbit secondary antibody After reacting at room temperature for 2 hours, store at -20°C.
[0040] 1) Investigate the activity of mouse secondary antibody after synthesis of mouse proximity ligation probe
[0041] Take 50 μl of mouse primary antibody with a concentration of 1ng / μl and coat it in a 96-well plate. After blocking, add 96 The well plate was combin...
Embodiment 2
[0049] Comparison of ELISA and ELISA-PLA to detect the effect of green fluorescent protein (GFP) in different sample doping
[0050] 1) Doping of green fluorescent protein in serum
[0051] ELISA experimental procedure: 50ng of mouse anti-GFP antibody was added to 50 μl of carbonate coating solution with pH 9.6 at 37°C for 2 hours, and then 1% bovine serum albumin was added at 37°C for 1 hour. Add 50 μl of the solution to be detected, incubate at 37 degrees Celsius for 1 hour, and wash three times with 20 mM phosphate buffer solution (pH 7.4), five minutes each time. After 50 ng of rabbit anti-GFP antibody was washed in 50 μl of 1% bovine serum albumin at 4°C for 12 hours, it was washed three times with 20 mM phosphate buffer solution (pH 7.4), five minutes each time. Add 50 μl of 1:5000 diluted horseradish peroxidase-labeled rabbit secondary antibody at 37 degrees Celsius for 1 hour, wash three times and develop color, the result is as follows Figure 5 as shown in (a);
...
Embodiment 3
[0057] Example 3 uses ELISA-PLA to detect phosphorylated proteins
[0058] 1) Site-specific phosphorylation antibody experimental group
[0059] ELISA-PLA experimental steps: 1:1000 dilution of mouse anti-ERK1 / 2 capture antibody in 50 μl of pH 9.6 carbonate coating solution at 37 degrees Celsius for 2 hours, then add 1% bovine serum albumin at 37 degrees Celsius for 1 hour . 1 μl of rat brain tissue extract was diluted to 50 μl, incubated at 37°C for 1 hour, and then washed three times with 20mM phosphate buffer solution (pH 7.4), five minutes each time. After adding 50 μl of 1:1000 diluted rabbit anti-phosphorylated ERK1 / 2 antibody to a 96-well plate, after 12 hours at 4°C, wash three times with 20 mM phosphate buffer solution (pH 7.4), five minutes each time. Other experimental steps are the same as above ELISA-PLA, the results are as follows Figure 6 As shown in (a), the phosphorylation level of ERK1 / 2 in chronic pain rats is higher than that in normal rats;
[0060] 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com