Method of regulating and controlling in-site catalytic polymerization reaction with bioenzyme on nano material on the basis of DNA
A technology for catalytic polymerization and nanomaterials, applied in the field of fixed-site catalytic polymerization, can solve the problems of no electroactivity, difficult to control the enzymatic polymerization reaction at a fixed point, hinder the formation of ordered polymers, etc., to achieve rapid reaction, low toxicity, biological The effect of high safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
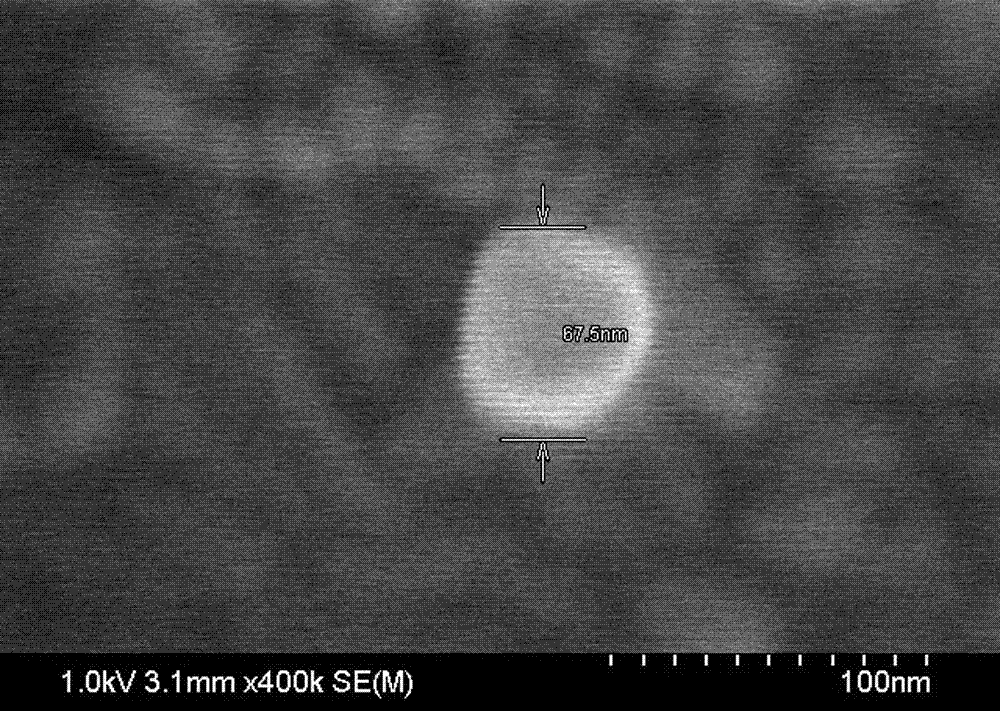
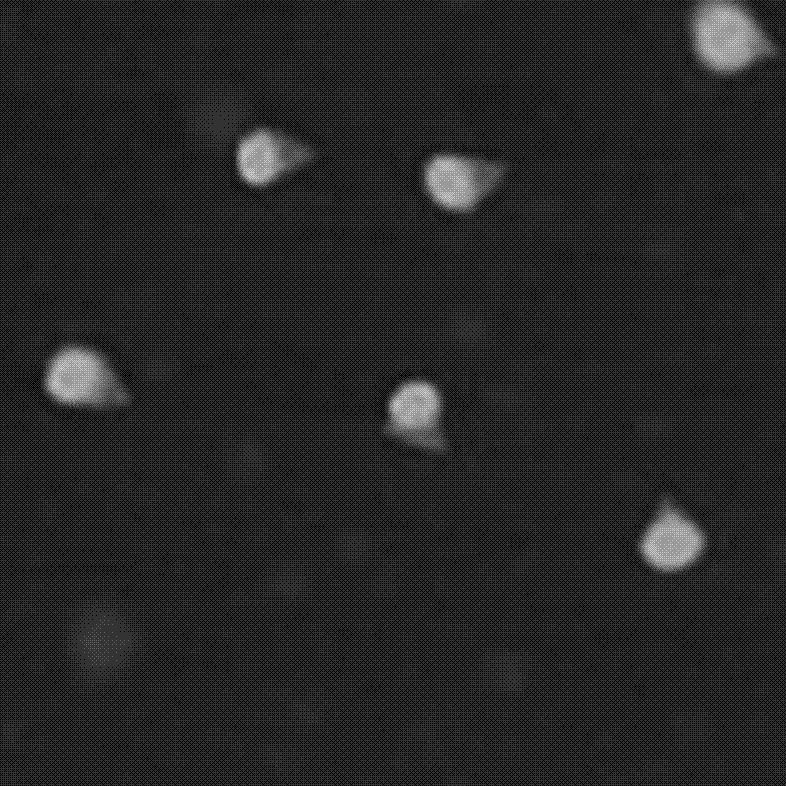
[0029] Mix 400 μL of 50 nm spherical gold nanoparticles (Au NPs, 1 nM) with single-stranded thiol-modified DNA 1 at a ratio of 1:3000, and slowly add sodium chloride (NaCl) and phosphate buffer (PB) to make Their final concentrations reached 0.1 M and 10 mM, respectively. After the mixture was incubated at 25°C for 8 hr, excess DNA was washed by centrifugation three times with 10 mM phosphate buffered saline (PBS). The resulting pellet was resuspended in PBS, biotin-modified single-stranded DNA 2 was added at a ratio of 1:3000, and incubated at 25°C to hybridize DNA 1 and DNA 2. After reacting for 8 hrs, centrifuge three times and use PBS to wash off excess DNA 2. After the obtained precipitate is resuspended in PBS, add 2 μL of 1 mM avidin-HRP to incubate for 30 min, centrifuge three times to wash off excess avidin-HRP, and use 400 Resuspend the pellet in μLPBS. Take 50 μL of the 200-fold diluted resuspension solution and add it dropwise on the cleaned conductive glass slid...
Embodiment 2
[0032] Design the DNA origami structure (the original sequence of the origami staple chain is the DNA sequence 14-221), DNA origami B-56 position protrudes a single strand-DNA hybridizes with the sulfhydryl-modified complementary strand DNA 3, at A-28, B-28, C-28 (respectively DNA 4, 5, 6) positions protrude single-stranded DNA. Mix 2 nM long-strand M13 DNA with 20 nM staple-strand DNA (including the replacement strand) in 1×TAE-Mg 2+ Mix well in buffer (40 mM tris(hydroxymethyl)aminomethane, 2 mM acetic acid, 2 mM disodium edetate, 12.5 mM magnesium acetate, pH 8.0), incubate at 95°C for 5 min, and slowly anneal to Centrifuge three times at 25°C with a 100 kd ultrafiltration tube to filter out excess single strands, recover the triangular origami structure in the tube, and quantify its concentration by ultraviolet light. HRP and SPDP were incubated at a ratio of 1:20 for 2 hr (PBS, pH=8.5), and ultrafiltered three times to wash away excess SPDP. Subsequently, HRP was incu...
Embodiment 3
[0035] Mix rod-shaped gold nanoparticles (Au NRs) and sulfhydryl-modified DNA 7 at a ratio of 1:3000, and slowly add sodium chloride (NaCl) and phosphate buffer (PB) to make the final concentrations of 0.1 M and 10 M, respectively. mM. After the mixture was incubated at 25°C for 8 hr, excess DNA was washed by centrifugation three times with 10 mM PBS. Design the DNA origami structure (the original sequence of the origami staple chain is DNA sequence 14-221), DNA origami B-56 position protrudes a single-stranded DNA to hybridize with the biotin-modified complementary strand DNA 5, and at the same time at B-45 , B-37, B-08, B-16, B-23 (DNA 8, 9, 10, 11, 12, respectively) stretch out the arm strands to capture Au NRs modified with DNA 4. Mix 2 nM long-strand M13 DNA with 20 nM staple-strand DNA in 1×TAE-Mg 2+Mix well in the buffer, incubate at 95°C for 5 minutes, anneal to 25°C, and filter out excess single strands with a 100 kd ultrafiltration tube. 5 nM of the obtained DNA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com