Low-abundance gene mutation detecting method
A gene and point mutation technology, which is used in the detection of low-abundance gene mutations, can solve the problems of reducing the efficiency of primer hybridization, affecting the competition between primers and repressors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
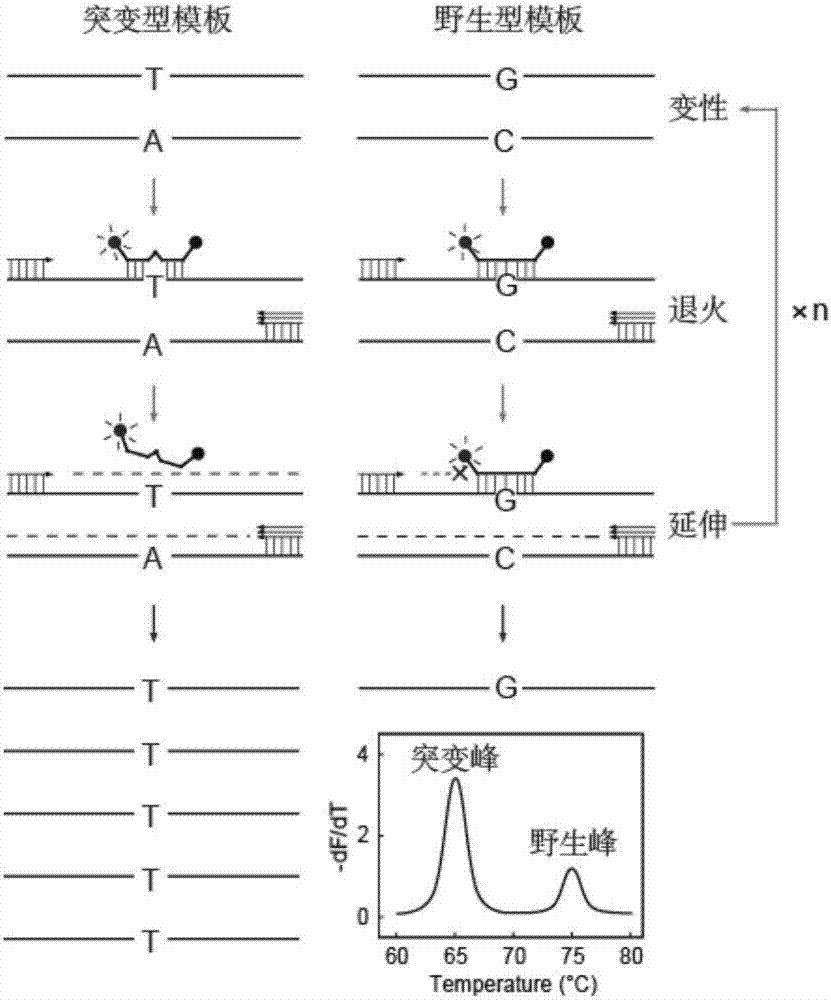
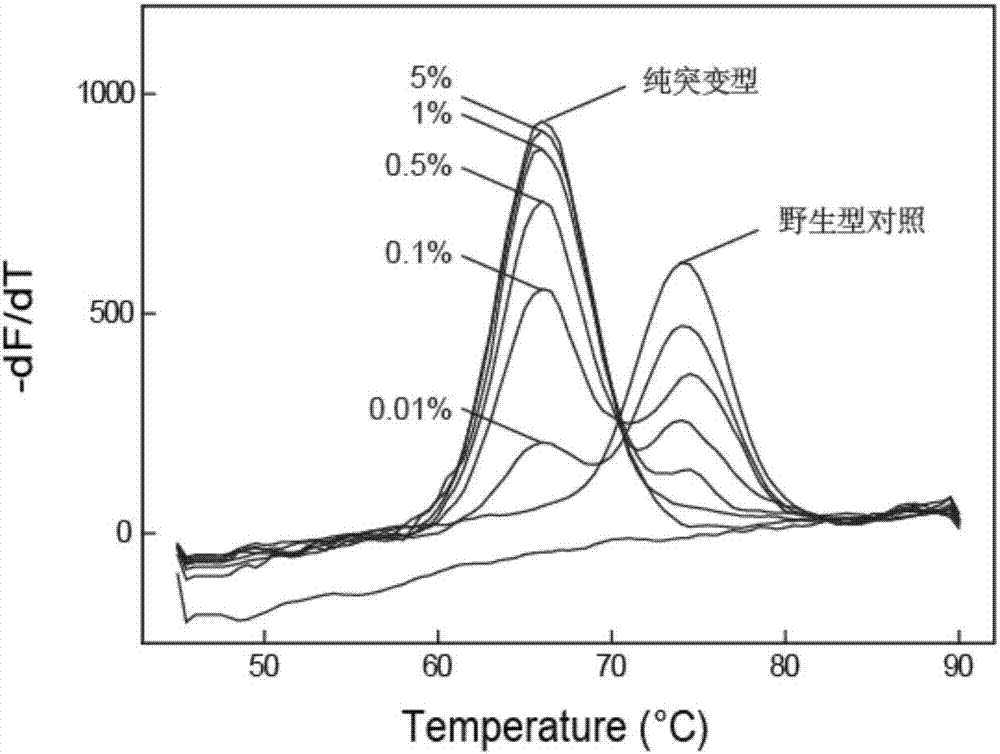
[0129] Example 1. Investigation of the ability of the self-quenching probe melting curve method that hinders primer extension to detect low-abundance mutations
[0130] The 315th codon of the catalase encoding gene (katG) of Mycobacterium tuberculosis is mutated from serine (Ser, S) to threonine (Thr, T) (AGC→ACC), which will lead to the first-line Resistance to the drug isoniazid (INH). In this example, taking the point mutation (katG S315T(AGC→ACC)) as an example, the ability of the self-quenching probe melting curve method that hinders primer extension to detect low-abundance mutations was investigated.
[0131] 1.1 Synthesis of primers and probes
[0132] In this example, an LNA-modified self-quenching probe and primer set for the katG gene were designed to ensure that the amplicon contained the mutation site. The forward primer is F1, the reverse primer is R1, and the probe is p1. Both the primer and probe sequences were synthesized by Shanghai Sangong, the primers wer...
Embodiment 2
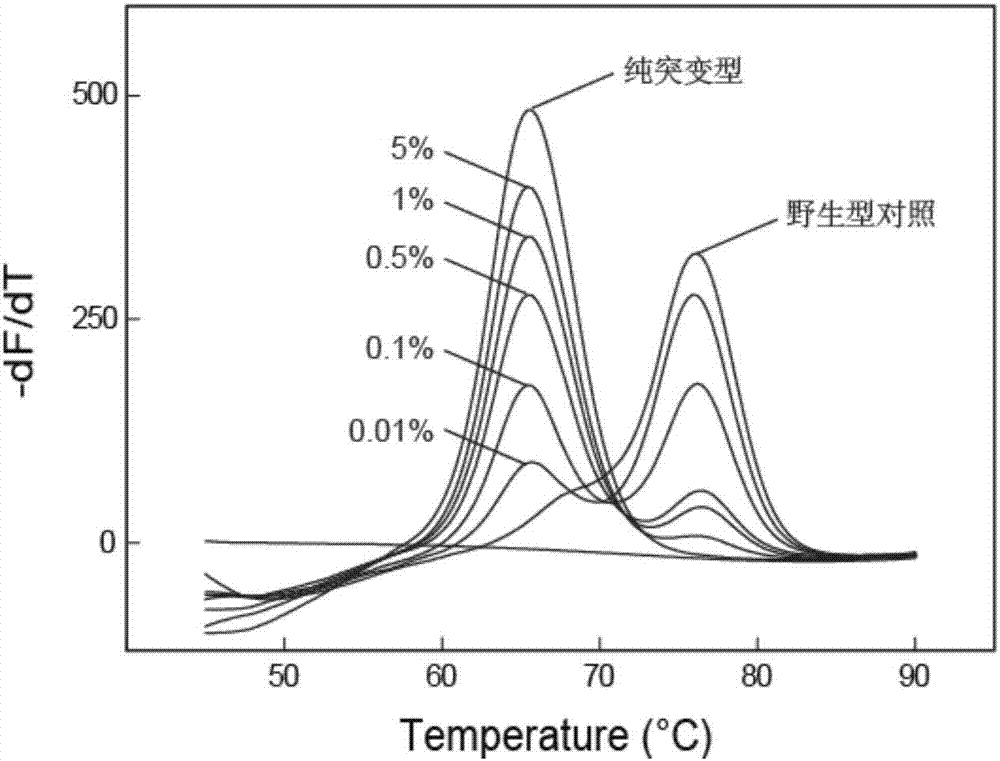
[0160] Example 2. Investigation of the ability of the self-quenching probe melting curve method that hinders primer extension to detect low-abundance mutations
[0161] In this example, taking the point mutation -15C→T in the promoter region of the inhA gene of Mycobacterium tuberculosis as an example, the ability of the self-quenching probe melting curve method that hinders primer extension to detect low-abundance mutations was investigated.
[0162] 2.1 Synthesis of primers and probes
[0163] In this example, an LNA-modified self-quenching probe and primer set targeting the promoter region of the inhA gene were designed to ensure that the amplicon contained the mutation site. The forward primer is F2, the reverse primer is R2, and the probe is p2. Both the primer and probe sequences were synthesized by Shanghai Sangong, the primers were purified by ULTRAPAGE, and the probes were purified by HPLC. The 5' end of the probe is labeled with ROX, and the 3' end is labeled with ...
Embodiment 3
[0190] Example 3. Investigation of the ability of the self-quenching probe melting curve method that hinders primer annealing to detect low-abundance mutations
[0191] In this example, taking the same point mutation (katG S315T (AGC→ACC)) as in Example 1 as an example, the ability of the self-quenching probe melting curve method that hinders primer annealing to detect low-abundance mutations was investigated.
[0192] 3.1 Design of primers and probes
[0193] First, design primer pairs and probes based on its gene sequence to ensure that the amplicon contains the mutation site. The forward primer is F2, and the reverse primer (R1) and self-quenching probe (p1) in this example are the same as those in Example 1. Wherein, the forward primer F2 and the probe have an overlapping sequence, so F2 and the probe can competitively bind to the region to be detected of the nucleic acid sequence to be detected, and when there is no mutation in the region, the probe binds to the target s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com