High-specificity microRNA fluorescence detection method based on short chain nucleic acid probe and double-chain specific endonuclease
An endonuclease and nucleic acid probe technology, which is used in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of reducing the overall speed of hybridization-enzyme cleavage reaction, slow miRNA hybridization, and difficulty in distinguishing single bases Mismatch and other problems, to achieve the effect of improving sensitivity, low cost, and high specificity constant temperature detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
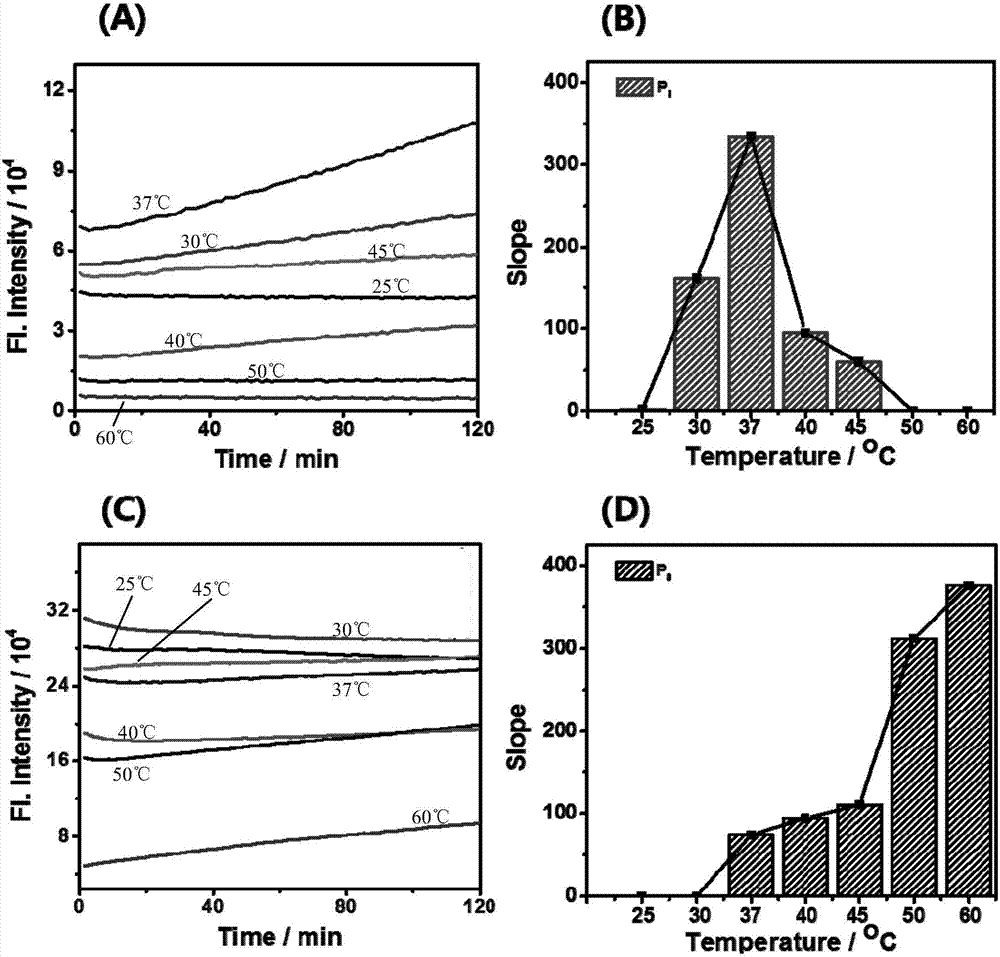
[0065] Embodiment 1 A kind of microRNA (hereinafter referred to as miRNA) fluorescent detection method based on short-chain nucleic acid probe and double-strand specific endonuclease
[0066] Design a short-chain DNA probe, which is labeled with a fluorescent group and a quencher group at both ends. Add the solution containing DNA probe and DSN enzyme to the solution to be tested. When there is no target miRNA in the solution to be tested, the fluorescent group labeled on the DNA probe is quenched and there is no fluorescence emission; when the solution contains target miRNA, The DNA probe hybridizes with the target miRNA, and the DSN enzyme cleaves the DNA probe in the DNA / miRNA duplex to release a fluorescent signal. The hybridization-enzyme digestion reaction occurs in a continuous cycle, and the fluorescent signal continues to increase. By optimizing the reaction conditions, a calibration curve for the detection of the target miRNA is obtained to achieve quantitative dete...
Embodiment 2
[0091] Embodiment 2 A kind of fluorescence detection method based on the microRNA let-7 family of short-chain nucleic acid probe and double-strand specific endonuclease
[0092] 1) Divide the target microRNA sequence into two sections, the 3' end section and the 5' end section, the base number of both sections is not less than 8bp, and the probe P 1 Make it possible to detect the 3' end segment or the 5' end segment;
[0093] Probe P 1 The number of bases is not less than 8bp, and does not exceed the base number of the 3' end segment or the 5' end segment; probe P 1 With fluorescent groups and quenching groups.
[0094] 2) with probe P 1 The microRNA sample to be tested is subjected to double-strand specific endonuclease-mediated microRNA fluorescence detection, and the fluorescence value F 1 , if with probe P 1 Complementary paired sequences only exist in the target microRNA, according to the fluorescence value F obtained by detection 1 Calculate the microRNA content, w...
Embodiment 3
[0099] Example 3 A microRNA let-7a fluorescence detection method based on short-chain nucleic acid probe and double-strand specific endonuclease
[0100] In this embodiment, the method of the present invention is further described by taking the detection of microRNA let-7a: 5'-UGA GGU AGU AGGUUG UAU AGUU-3' (SEQ ID NO: 4) in the microRNA let-7 family as an example.
[0101] (1) Divide the target microRNA let-7a sequence into two segments, the 3' end segment and the 5' end segment, and detect the probe 7a-P of the 3' end segment according to the design 1 ; Simultaneously design a probe 7a–P that is completely complementary to the entire sequence of microRNA let-7a 0 As a control, all probes are labeled with fluorescent groups and quenchers, and the specific sequences are as follows:
[0102] Probe 7a-P 1 : FAM-5'-CTATACAACC-3'-BHQ-1 (SEQ ID NO: 1),
[0103] Probe 7a–P 0 : FAM-5'-AACTATACAACCTACTACCTCA-3'-BHQ-1 (SEQ ID NO: 3).
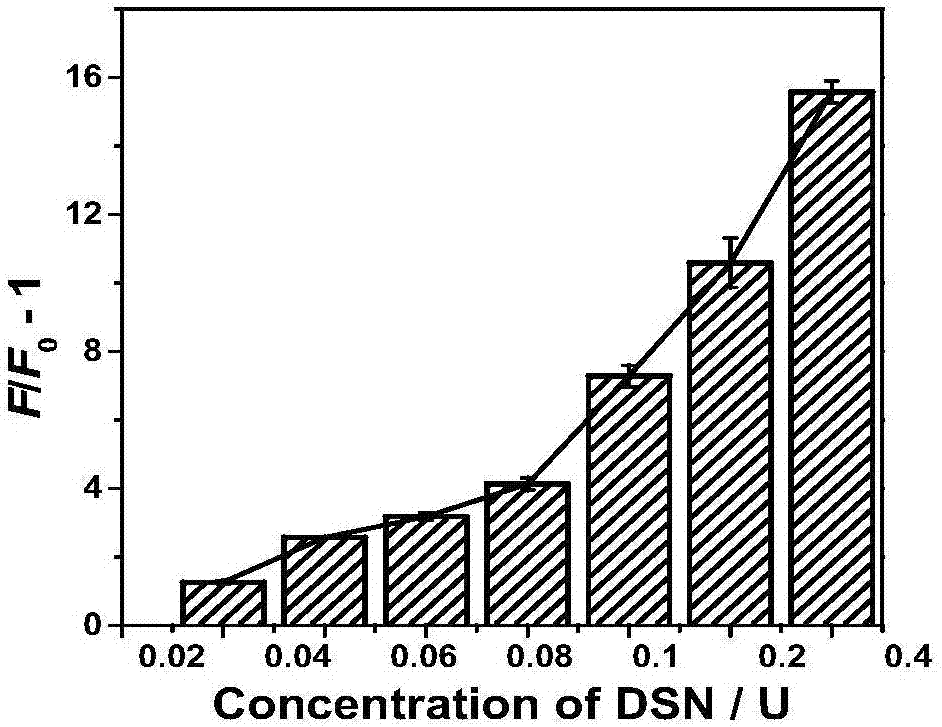
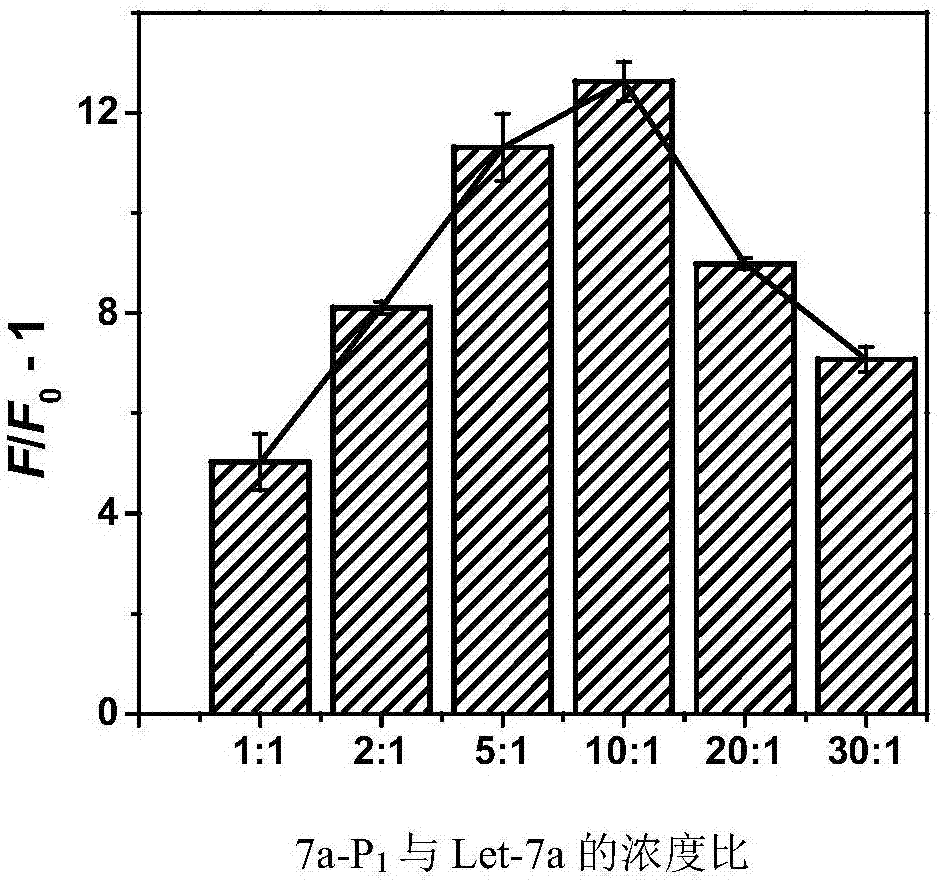
[0104] (2) Probe 7a-P 1 ,7a–P 0 Optimizatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com