Method for rapid high-efficiency qualitative and quantitative analysis of N-sugar in virtue of high-performance liquid chromatography
A high-performance liquid chromatography and quantitative analysis technology, which is applied in the direction of analyzing materials, measuring devices, and material separation, can solve the problems of unsatisfactory sensitivity and long time consumption, so as to improve analysis efficiency and detection sensitivity, enhance analysis sensitivity, and respond The mild effect of the conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] A fast and efficient N-sugar qualitative and quantitative analysis method utilizing high performance liquid chromatography, specifically comprising the following steps:
[0025] (1) Add 10 μg of glycoprotein RNase B, fetuin or 10 μL of human serum to ammonium bicarbonate buffer and denaturing buffer at pH 8.5. The total volume is 20 μL, of which the denaturing buffer is 0.48 μL. Denatured in a boiling water bath for 10 minutes, after cooling to room temperature, 2.4 μL of 10% NP-40 was added to inhibit the denaturant for 5 minutes.
[0026] (2) Add 1 μL of exoglycosidase PNGse F, microwave-assisted enzyme digestion for 20 min, in order to prevent the sample from boiling, the sample was floated in a beaker filled with 500 mL of water and reacted in a microwave oven with 700W power.
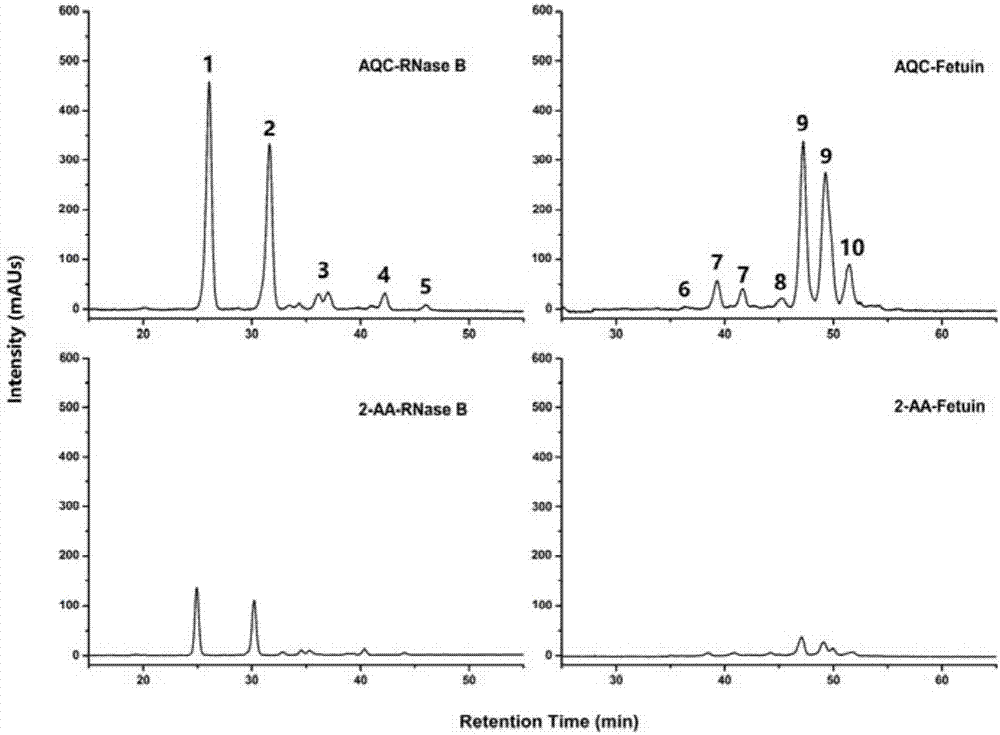
[0027] (3) After the reaction is completed, cool to room temperature, then directly add 20 μL of 150 mM AQC acetonitrile solution and 20 μL of pH 8.5 ammonium bicarbonate buffer, shake and m...
Embodiment 2
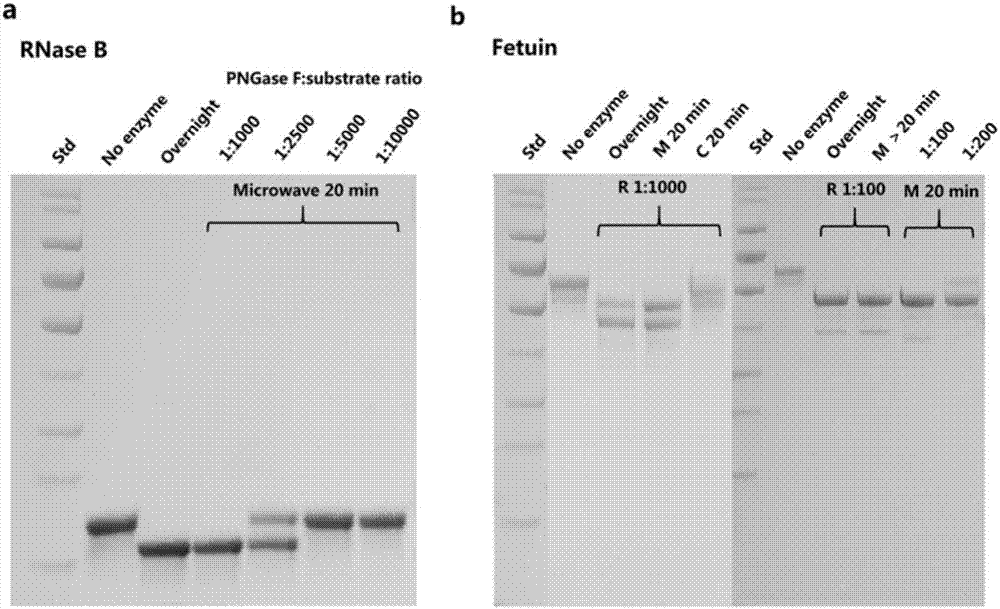
[0030] Glycoprotein RNase B was selected for condition optimization, fetuin was used for further verification, and protein SDS-PAGE was used to explore the effects of microwave time and the molar ratio of enzyme to substrate during PNGase F digestion, and compared with the traditional overnight digestion that can completely digest By comparison, the most suitable molar ratio of enzyme and substrate and microwave irradiation time were obtained. Specific steps are as follows:
[0031] (1) After 10 μg of RNase B is denatured in the manner of Example 1;
[0032] (2) Control the microwave radiation time to 20min, add 1:10000, 1:5000, 1:2500, 1:1000 molar ratio of enzyme to substrate for experiment, and carry out the experiment with the traditional overnight enzyme digestion that can completely digest Compared;
[0033] (3) Select the optimized molar ratio of enzyme and substrate in step (2) of 1:1000, and do a verification test on the complex glycoprotein fetuin, and find that th...
Embodiment 3
[0037] After RNase B digestion was completed using the optimized microwave-assisted digestion conditions, 20 μL of AQC solution and 20 μL of digestion reaction buffer were added, and the reaction was performed in a water bath.
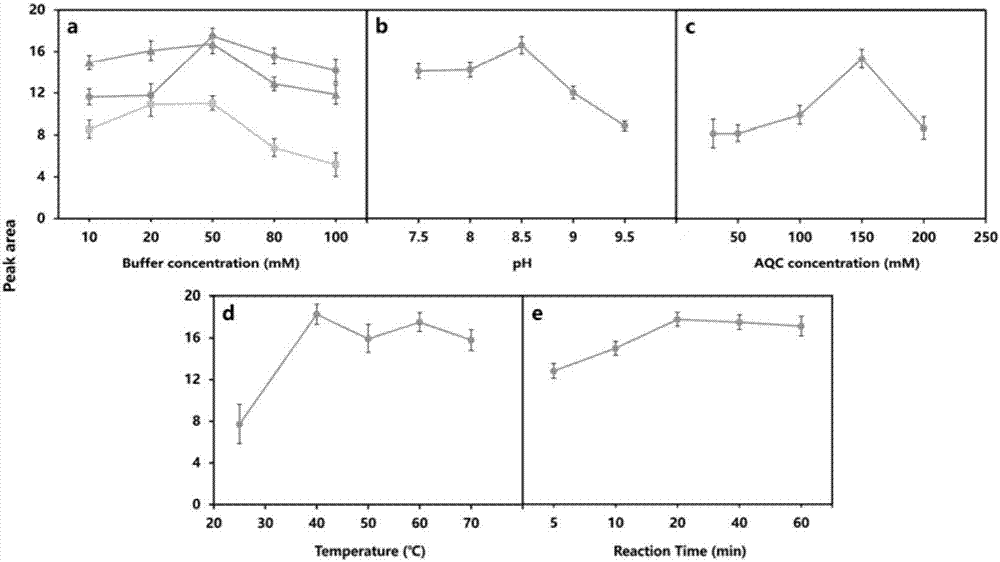
[0038] Through the factors affecting AQC labeling reaction, the different types of reaction buffer (sodium phosphate buffer, ammonium bicarbonate buffer, sodium borate buffer) and concentration (10, 20, 50, 80, 100) mM, the concentration of derivative reagent (30, 50, 100, 150, 200) mM, reaction pH (7.5, 8.0, 8.5, 9.0), temperature (25, 40, 50, 60, 70) °C and reaction time (5, 10, 20, 40 , 60) min for exploration and optimization. The optimization experiment adopts a single factor analysis method, that is, one factor is selected for gradient analysis, and other factors are kept the same. After confirming the condition of one factor, the analysis of other factors is carried out similarly.
[0039] attached by figure 2 The optimal reaction condition ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com