Nosema pernyi template DNA extraction method and application of nosema pernyi template DNA extraction method to molecular diagnosis
A technology for microsporidia and molecular diagnosis, applied in microorganism-based methods, DNA preparation, recombinant DNA technology, etc., can solve the problems of low detection sensitivity, cumbersome operation, low throughput, etc., and achieve high throughput and interference factors. Less, overcome the effect of tissue blackening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
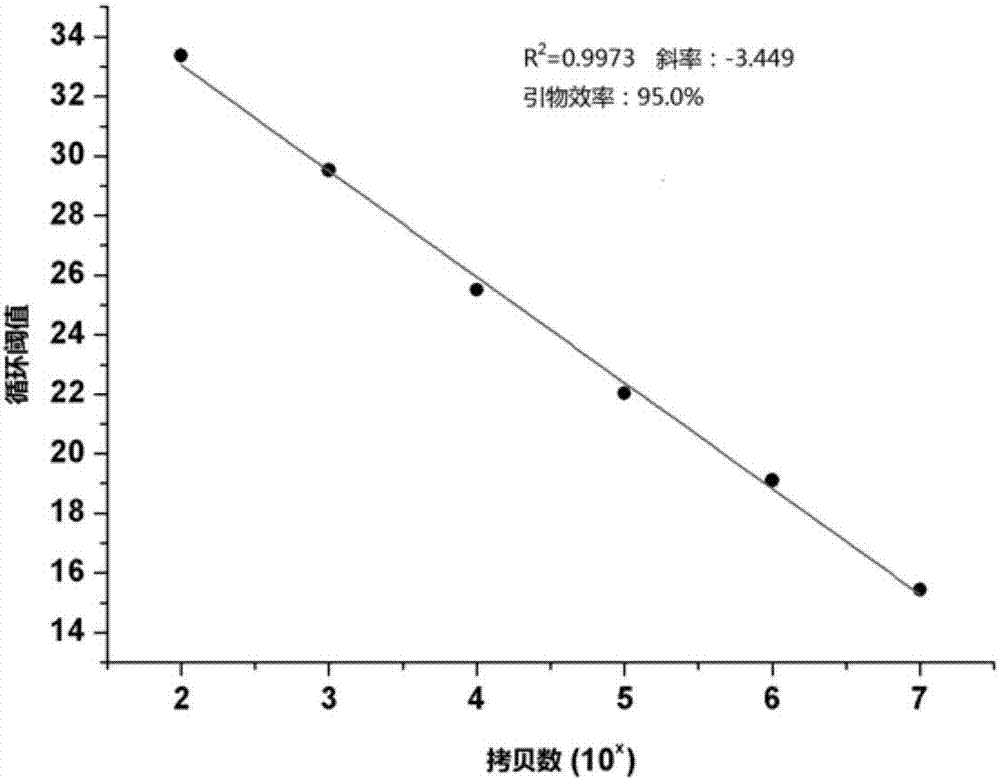
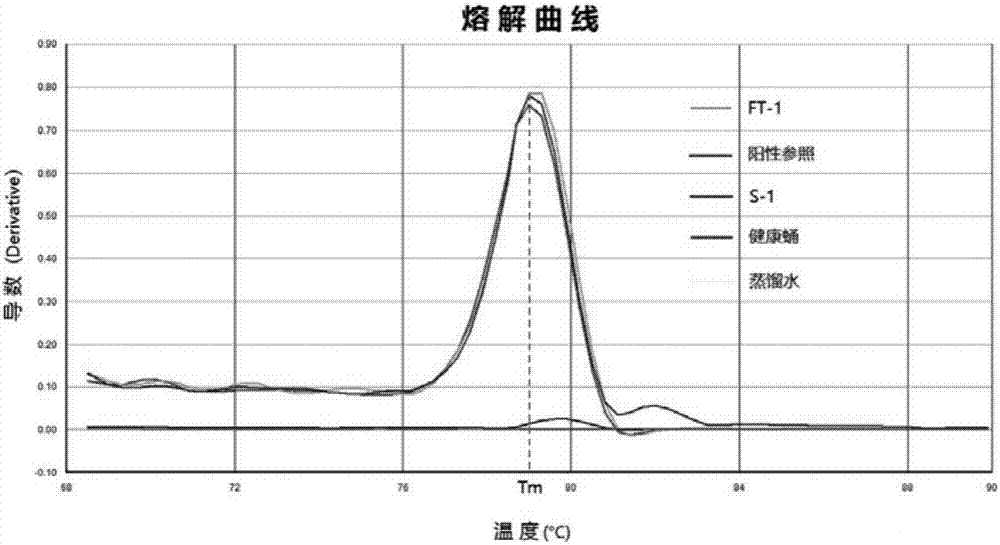
[0062] Acquisition of template DNA for No. tussah mori and diagnosis of No. tussah by SYBR Green fluorescence quantitative PCR
[0063] Select a diseased pupa infected with No. tussah mori, and put a small amount of tissue into two 1.5mL EP tubes, numbered S-1 and FT-1 respectively. Add 200 μL of lysate S (0.2M NaOH, 1% SDS) to tube S-1, oscillate to mix, let it stand at room temperature for 40 minutes, add magnetic bead suspension 10 μL (sigma) and absolute ethanol 150mL, oscillate gently to mix , place the EP tube on a magnetic stand for separation, and when the magnetic beads are completely separated from the liquid phase, pour off the liquid phase, remove the magnetic stand, add 150 μL of 70% ethanol, oscillate and mix well, place on the magnetic stand for separation, discard For liquid, remove the magnetic stand, add 150 μL of 70% ethanol again, oscillate, mix and place on the magnetic stand for separation, pipette the liquid as much as possible, remove the magnetic stand...
Embodiment 2
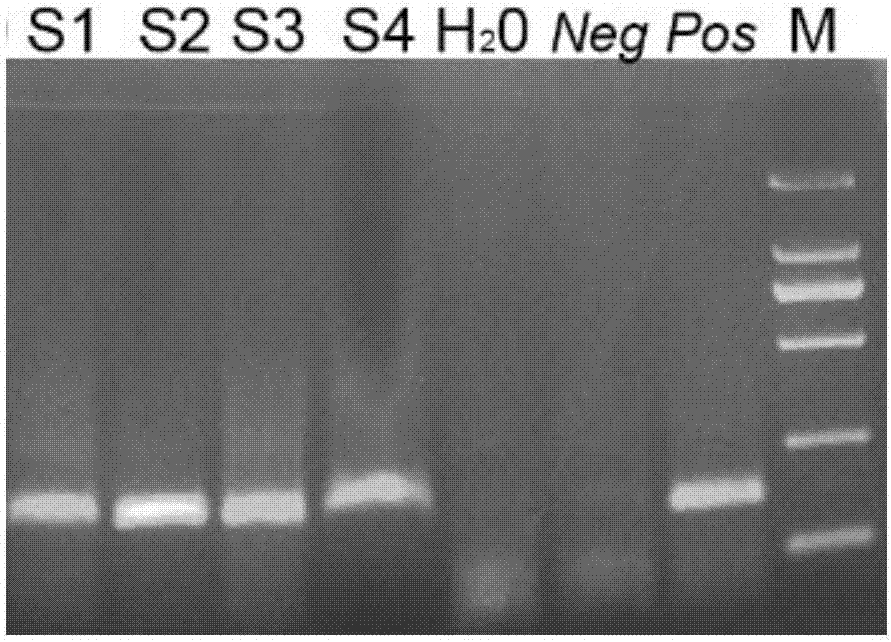
[0071] Acquisition of template DNA of No. tussah silkworm and routine PCR diagnosis of No. tussah silkworm.
[0072]Four diseased pupae infected with No. tussah silkworm were selected, numbered S1, S2, S3 and S4 respectively. A small amount of tissue was taken from S1 and S2 respectively. Among them, S1 used a scalpel to cut an incision of about 5 mm on the back of the pupae, quickly picked up a little tissue with a toothpick, and then disinfected the incision with 75% alcohol cotton. Make a wax seal. Put the S1 pupa into the cocoon shell, seal the cocoon shell with adhesive tape, place it at room temperature (15-30°C), and observe the hatching situation. Take a small amount of pupal skin and pupal blood mixture in S3, and take a small amount of pupal skin and tissue mixture in S4, and place them in four 1.5mL EP tubes respectively. Add 200 μL of lysate S (0.2M NaOH, 1% SDS) respectively, oscillate and mix well, leave at room temperature for half an hour, add 10 μL of magnet...
Embodiment 3
[0077] Choose 60 female moths that have laid eggs, numbered respectively 1#-60#. A small amount of tissue was picked and placed in 60 1.5mL EP tubes containing 200μL 0.2M KOH (among them, 4 samples were taken from 2 eggs each due to the small amount of tissue in the belly of the moth). Freeze the tubes at -20°C, remove and thaw after 30 days, transfer 100 μL of the lysate supernatant in each EP tube to different sample wells in a 96-well plate, add 10 μL of magnetic bead suspension (Sigma) and 75 mL of absolute ethanol , gently oscillate and mix, put the EP tube on a 96-well magnetic stand for separation, wait until the magnetic beads are completely separated from the liquid phase, pour off the liquid phase, remove the magnetic stand, add 100 μL of 70% ethanol each, oscillate and mix Then put it on the magnetic stand for separation, discard the liquid, remove the magnetic stand, add 100 μL of 70% ethanol again, oscillate and mix well, and place it on the magnetic stand for sep...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com