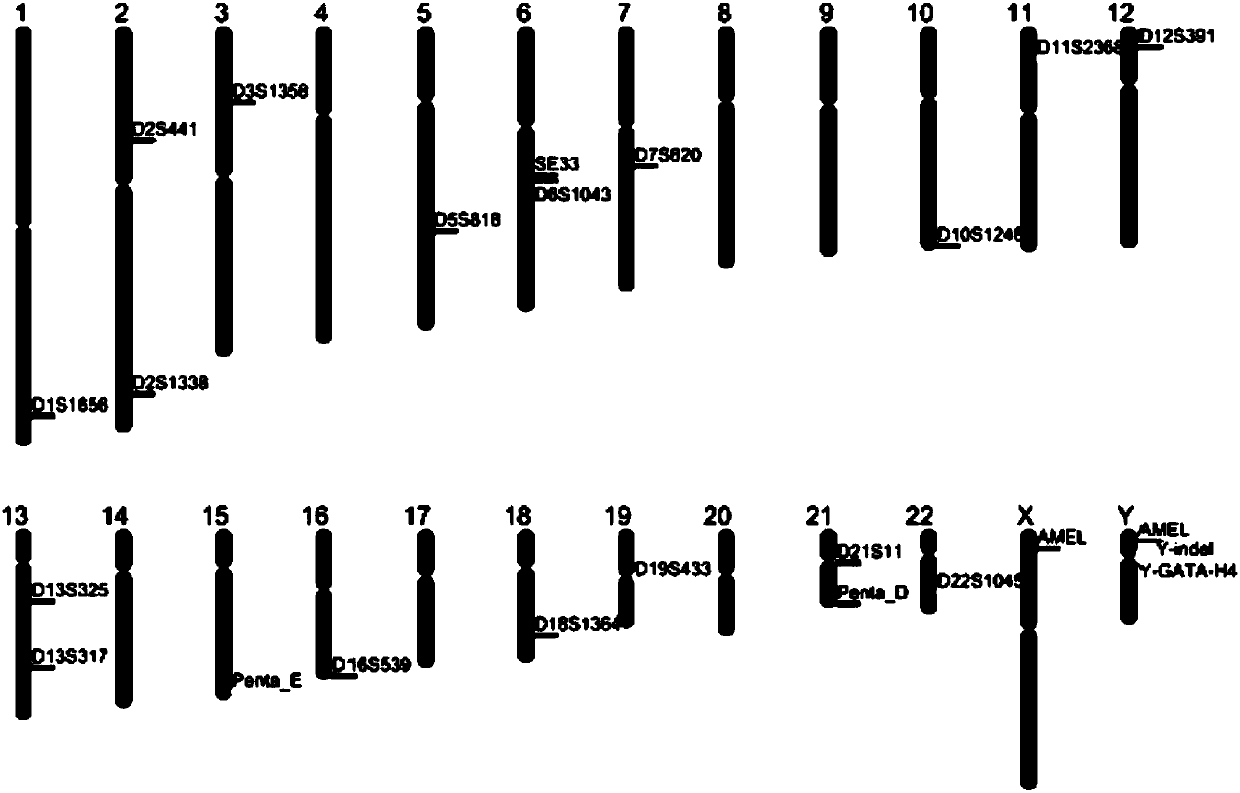
Complex amplification system applied to 23 human STR sites, kit and application of kit
A complex amplification system, D21S11 technology, applied in the biological field, achieves the effects of high accuracy, high polymorphism, and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Utilize the fluorescence-labeled composite amplification kit of 23 STR sites of the present invention to conduct blood samples from a doublet (father-son) and triplet (parent-daughter) family (the sample has been identified and confirmed by the Haiershi HGT 21G kit) to test.
[0076] The family blood samples were donated by volunteers, and the DNA was extracted by the chelex-100 method (for specific methods, refer to "Forensic DNA Protocol", Humana Press, 1998). Take 3 μL of blood with anticoagulants in a 1.5ml centrifuge tube, shake and mix the chelex Solution, to make chelex fully suspended, add 200μL 5% chelex-100 to each tube, shake the sample, after incubating on a constant temperature metal bath at 56°C for 2 hours, take out the sample and shake for 2 minutes, boil for 8-10 minutes, centrifuge at 13000rpm for 3 minutes, be careful Aspirate about 150 μL of supernatant and transfer to a new 1.5ml centrifuge tube. Using the kit of the present invention and Hirsch's ...
Embodiment 2
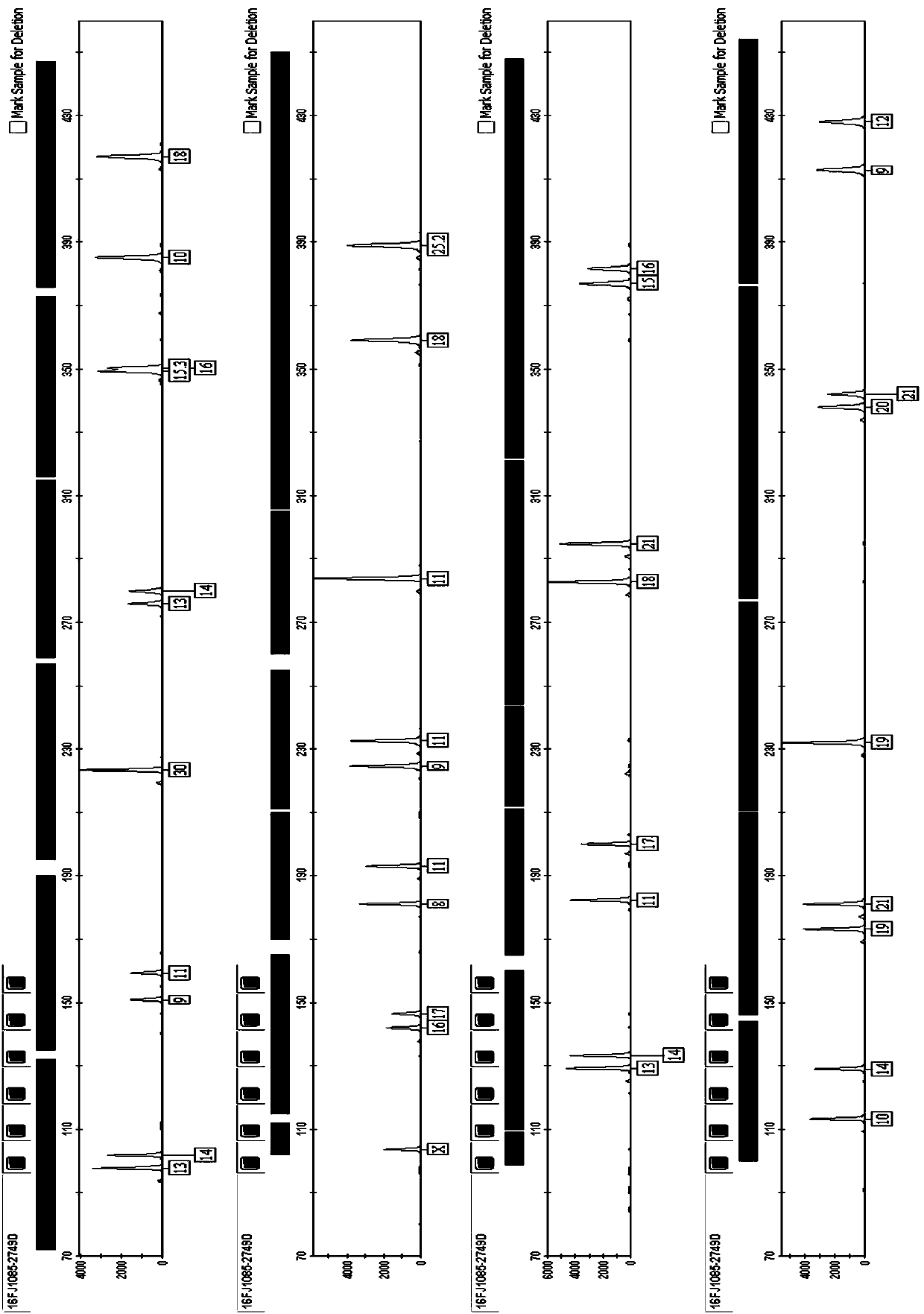
[0101] Multiple amplification and typing were performed on blood sample 1, where blood sample 1 was donated by volunteers, and the adjacent alleles of D1S1656 in blood sample 1 could not be correctly typed using the existing HGT 21G kit.
[0102] The DNA was extracted by the chelex-100 method (refer to "Forensic DNA Protocol" for specific methods, HumanaPress, 1998), and the sample was amplified and detected simultaneously using the kit of the present invention and the HGT 21G kit of Haiers Gene Technology Co., Ltd. The amplification reaction was carried out on the G1000 thermal cycler, the electrophoresis and detection were carried out on the ABI 3500 genetic analyzer, and the data analysis was carried out by Gene IDx software. The reagents and materials used, such as formamide, internal standard, etc., are conventional materials commonly used by those skilled in the art.
[0103] 2.1 DNA extraction by Chelex-100 method
[0104] 2.2 Polymerase chain reaction (PCR) amplific...
Embodiment 3
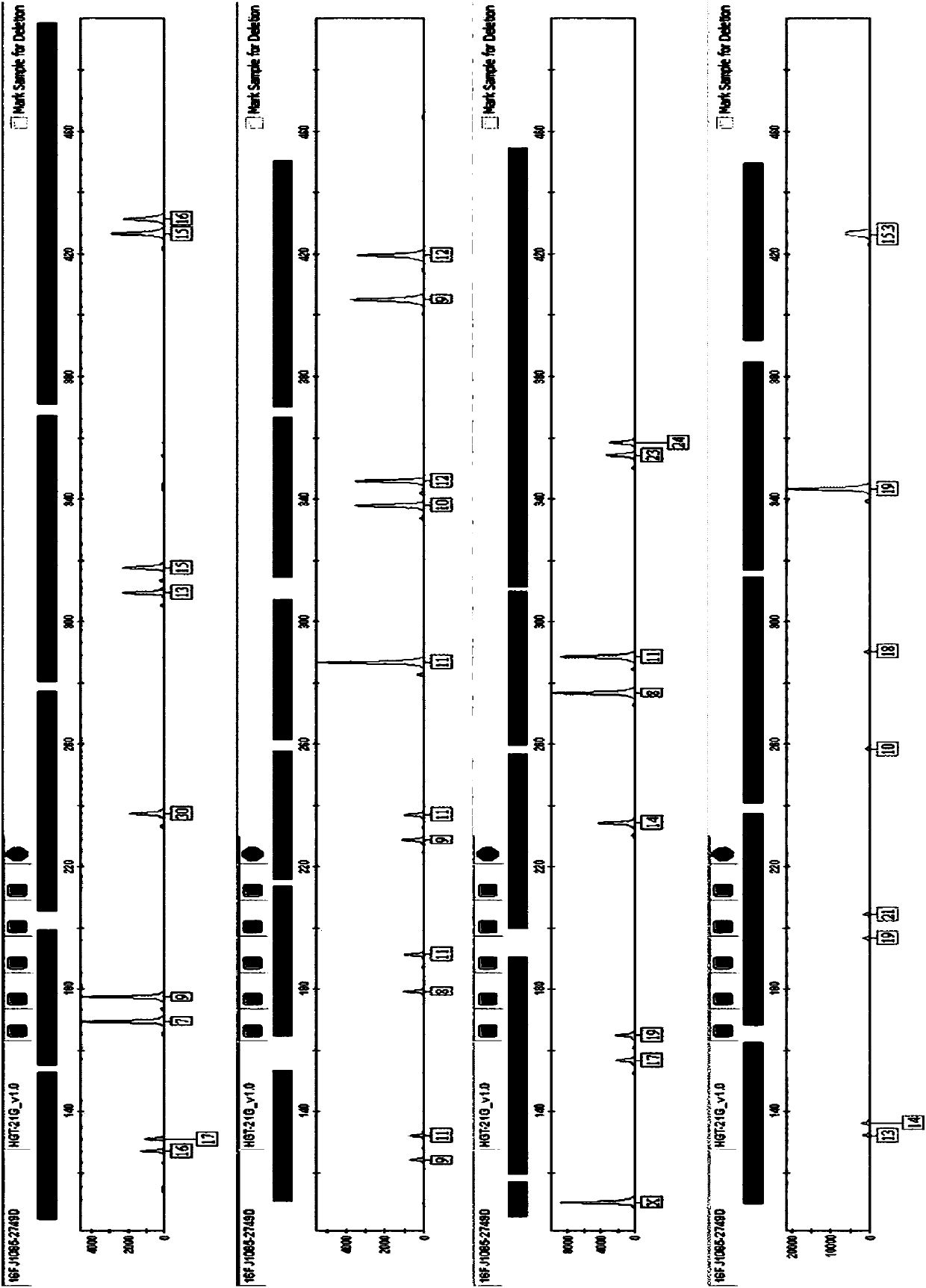
[0114] Multiplex amplification and typing were performed on the three loci of blood sample 2 (including sample A, sample B, and sample C). Blood sample 2 was donated by volunteers, and each blood sample 2 contained common microvariant alleles, which could not be correctly typed using the existing MR23sp kit from Yuewei Gene Company.
[0115] DNA adopts the chelex-100 method to extract (specific method with reference to " Forensic DNA Protocol ", HumanaPress, 1998), utilizes kit of the present invention, the kit MR23sp of Yuewei Gene Company to amplify above-mentioned sample 2 simultaneously, amplification reaction is heated in G1000 It was carried out on a cycler, electrophoresis and detection were carried out on an ABI 3500 genetic analyzer, and data analysis was performed using GeneMapperIDx software. The reagents and materials used, such as formamide, internal standard, etc., are conventional materials commonly used by those skilled in the art.
[0116] 3.1 DNA extraction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com