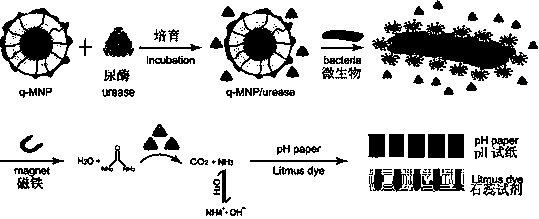
Platform rapid microorganism detection method based on quaternary ammonium salt magnetic nanometer material and urease composite sensor
A composite sensor, quaternary ammonium salting technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of complex operation, extremely high operator requirements, and long time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
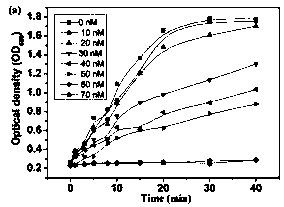
[0027] Example 1: Detection of Staphylococcus aureus.
[0028] The microorganisms used in the experiment were cultured in suspension in lysed broth (1% peptone, 1% sodium chloride, 0.5% yeast extract, 100 mL water), and a single colony was cultured overnight at 30°C and 200 rpm on a shaking table at 4500 rpm. / Centrifuge for ten minutes, and dilute to different concentrations with PBS buffer solution.
[0029] 100 nM urease solution was incubated with 6nM q-MNP1 for 15min, and 100 µL concentration (10 cfu ml -1 ) bacterial solution and the above-mentioned nanomaterial and enzyme complex, reacted at 37 °C for 30min. Use a magnet to remove magnetic particles or magnetic particle-microbe complexes, then add 200 mM NaCl, 60 mM MgCl 2 , 200 microliters of 50mM urea and 0.4% phenol red solution, incubate for 30 minutes, and quickly detect the microbial intensity signal with a multi-functional microplate reader.
[0030] 100 nM urease solution was incubated with 6nM q-MNP1 for 15...
Embodiment 2
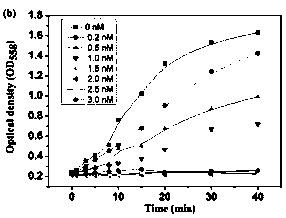
[0038] Example 2: Detection of Escherichia coli.
[0039] The microorganisms used in the experiment were cultured in suspension in lysed broth (1% peptone, 1% sodium chloride, 0.5% yeast extract, 100 mL water), and a single colony was cultured overnight at 30°C and 200 rpm on a shaking table at 4500 rpm. / Centrifuge for ten minutes, and dilute to different concentrations with PBS buffer solution.
[0040] 100 nM urease solution was incubated with 6nM q-MNP2 for 15min, and 100 µL concentration (10 cfu ml -1 ) bacterial solution and the above-mentioned nanomaterial and enzyme complex, reacted at 37 °C for 30min. Use a magnet to remove magnetic particles or magnetic particle-microbe complexes, then add 200 mM NaCl, 60 mM MgCl 2 , 200 microliters of 50mM urea and 0.4% phenol red solution, incubate for 30 minutes, and quickly detect the microbial intensity signal with a multi-functional microplate reader.
[0041] 100 nM urease solution was incubated with 6nM q-MNP2 for 15min, ...
Embodiment 3
[0049] Example 3: Detection of Edwardsiella tarda bacteria.
[0050] The microorganisms used in the experiment were cultured in suspension in lysed broth (1% peptone, 1% sodium chloride, 0.5% yeast extract, 100 mL water), and a single colony was cultured overnight at 30°C and 200 rpm on a shaking table at 4500 rpm. / Centrifuge for ten minutes, and dilute to different concentrations with PBS buffer solution.
[0051] 100 nM urease solution was incubated with 6nM q-MNP3 for 15min, and 100 µL concentration (10 cfu ml -1 ) bacterial solution and the above-mentioned nanomaterial and enzyme complex, reacted at 37 °C for 30min. Use a magnet to remove magnetic particles or magnetic particle-microbe complexes, then add 200 mM NaCl, 60 mM MgCl 2 , 200 microliters of 50mM urea and 0.4% phenol red solution, incubate for 30 minutes, and quickly detect the microbial intensity signal with a multi-functional microplate reader.
[0052] 100 nM urease solution was incubated with 6nM q-MNP3 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com