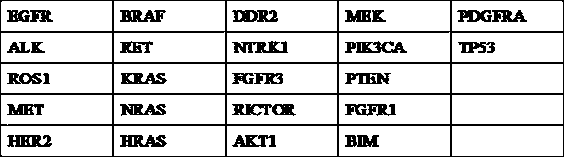
High-flux detection method of circulating tumor DNA lung cancer driver gene
A detection method, a high-throughput technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of patient suffering, invasiveness, and difficulty in obtaining patient samples, and achieve strong predictability and accuracy high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0044] A kind of detection method of circulating tumor DNA lung cancer gene of the present invention, comprises the steps:
[0045] 1. Sample library preparation:
[0046] (1) Pipette 60 ul of purified DNA (ctDNA) and 40 ul of Endprep mix into a single PCR tube, pipette to mix, and briefly centrifuge for end repair. The reaction conditions are 30°C and 30 min.
[0047] (2) Take out the magnetic beads half an hour in advance to return to room temperature, shake and mix, take 180ul of magnetic beads and add them to the end-repaired PCR product, beat and mix, and incubate in the greenhouse for 5 minutes.
[0048] (3) Put the PCR single tube on the magnetic stand, let it stand for 5 minutes, remove the supernatant, keep the PCR tube on the magnetic stand, add 200ul of 80% ethanol (newly prepared) to rinse, let stand for 30s, remove the supernatant, 80 Rinse twice with % ethanol, and dry at room temperature for 10 minutes (with 2 minutes remaining, use a small grab to suck up the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com