Nucleic acid aptamer for detecting human pdl1 protein and its application in preparation of detection preparation
A nucleic acid aptamer and human detection technology, applied in the field of molecular biology, can solve the problems of nucleic acid aptamer reports without human PDL1 protein, and achieve the effects of easy in vitro chemical synthesis, stable sequence and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
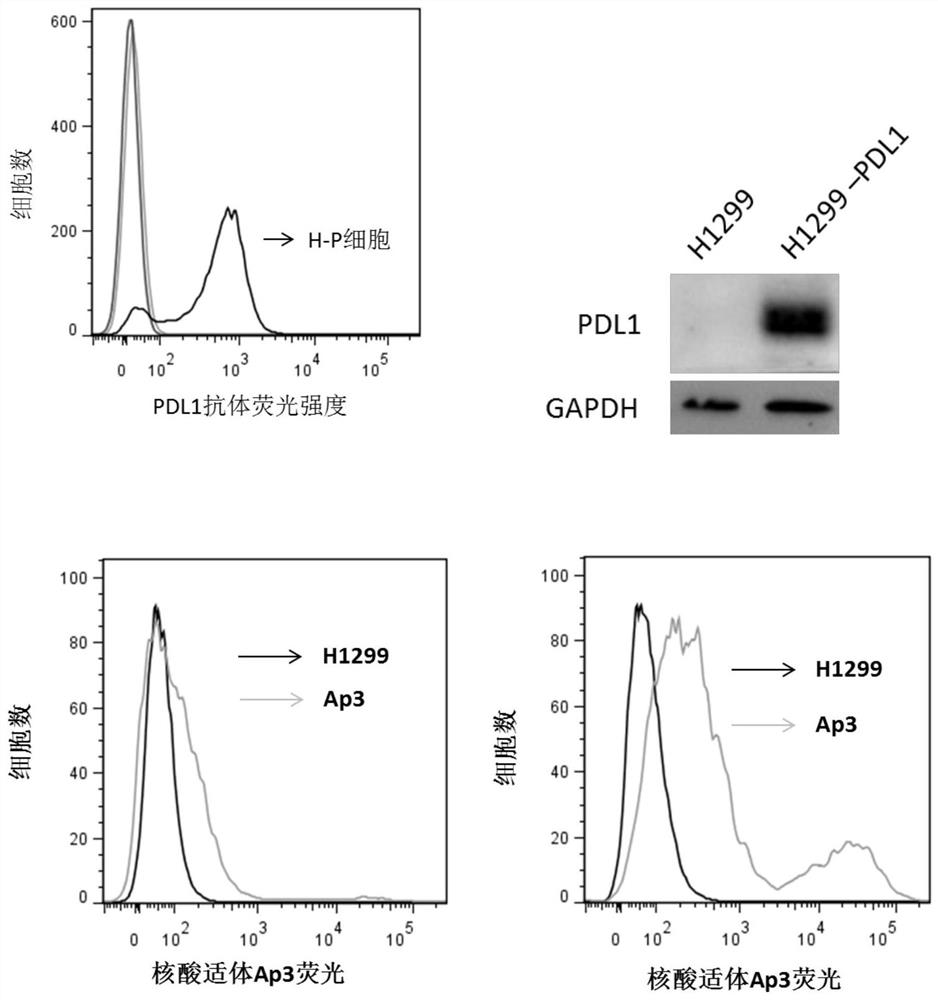
[0042] Embodiment 1: the screening of the aptamer of human PDL1 protein specific binding
[0043] The PDL1 plasmid was constructed, transfected into the non-small cell lung cancer cell line H1299, and the cell line with stable and high expression of PDL1 was screened by sorting flow cytometry and designated as H1299-PDL1(H-P). Using the aptamer library related to non-small cell lung cancer reported so far, we investigated its binding ability to H1299 cells and H1299 cells (H-P) overexpressing PDL1 protein, and finally selected an aptamer with obvious differences and named it Ap3.
Embodiment 2
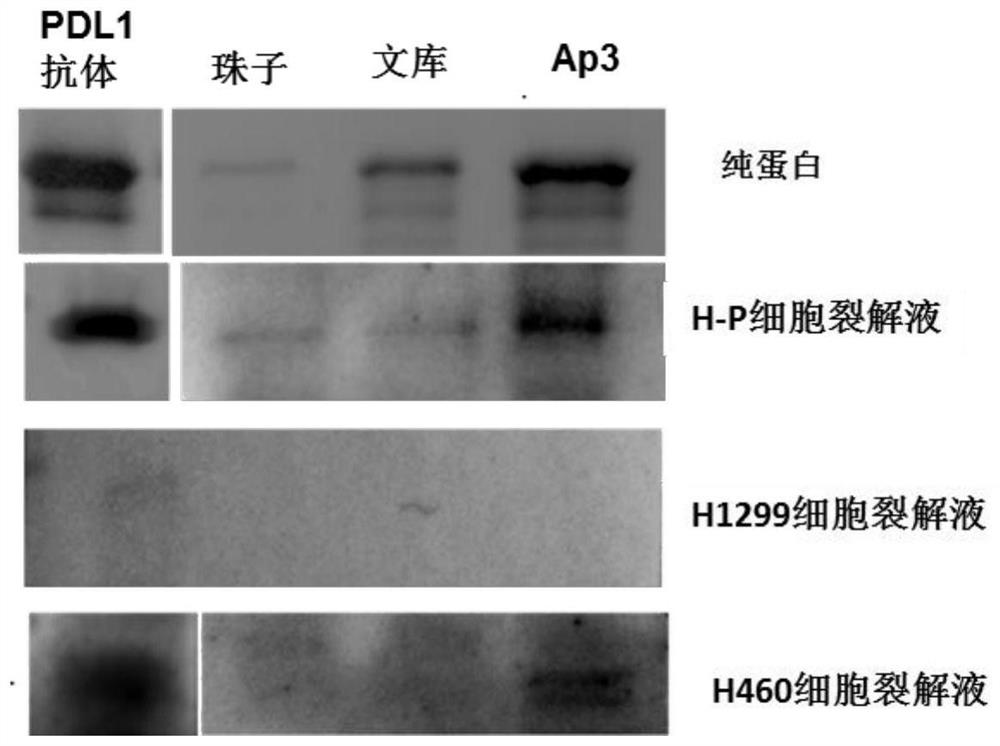
[0044] Example 2: Defining the binding of nucleic acid aptamer Ap3 to PDL1 protein
[0045] Incubate H-P cell lysate with aptamer Ap3 or library at 4°C for 1h, then incubate the mixture with streptavidin-labeled agarose beads at 4°C for 1h, wash with 2X protein loading buffer The protein bound to the beads was eluted and analyzed by Western Blotting.
[0046] Human pure PDL1 protein (Shenzhou Biological Technology Co., Ltd.) and Ap3 or library were incubated at 4 degrees for 1 hour, then the mixture was incubated with streptavidin-labeled agarose beads at 4 degrees for 1 hour, washed with 2X The proteins bound to the beads were eluted with protein loading buffer and analyzed by Western Blotting.
Embodiment 3
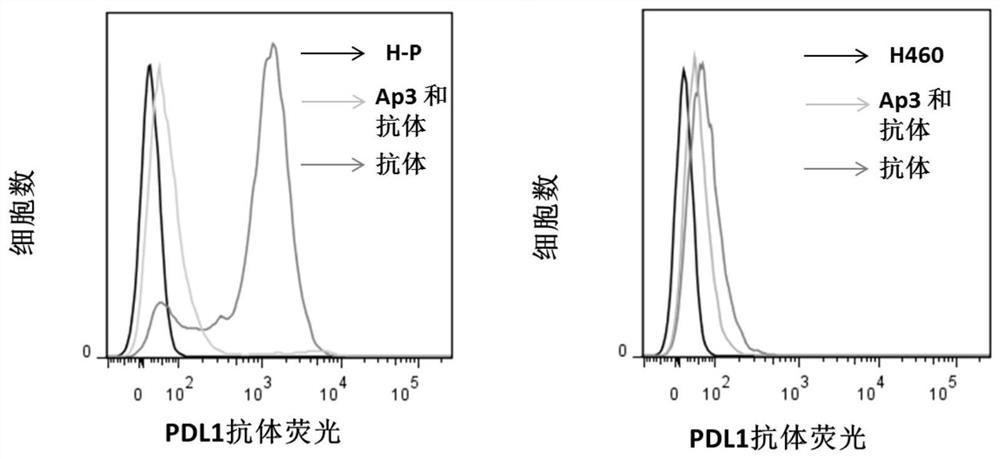
[0047] Example 3: Nucleic acid aptamer Ap3 can compete with PDL1 antibody for binding to H-P cells
[0048] The nuclear PDL1 antibody was incubated with H-P cells alone or Ap3 and PDL1 antibody 20 times higher than the PDL1 antibody were incubated with H-P cells at the same time. After washing, the fluorescence intensity (PE) of the PDL1 antibody bound to the cells was compared.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com