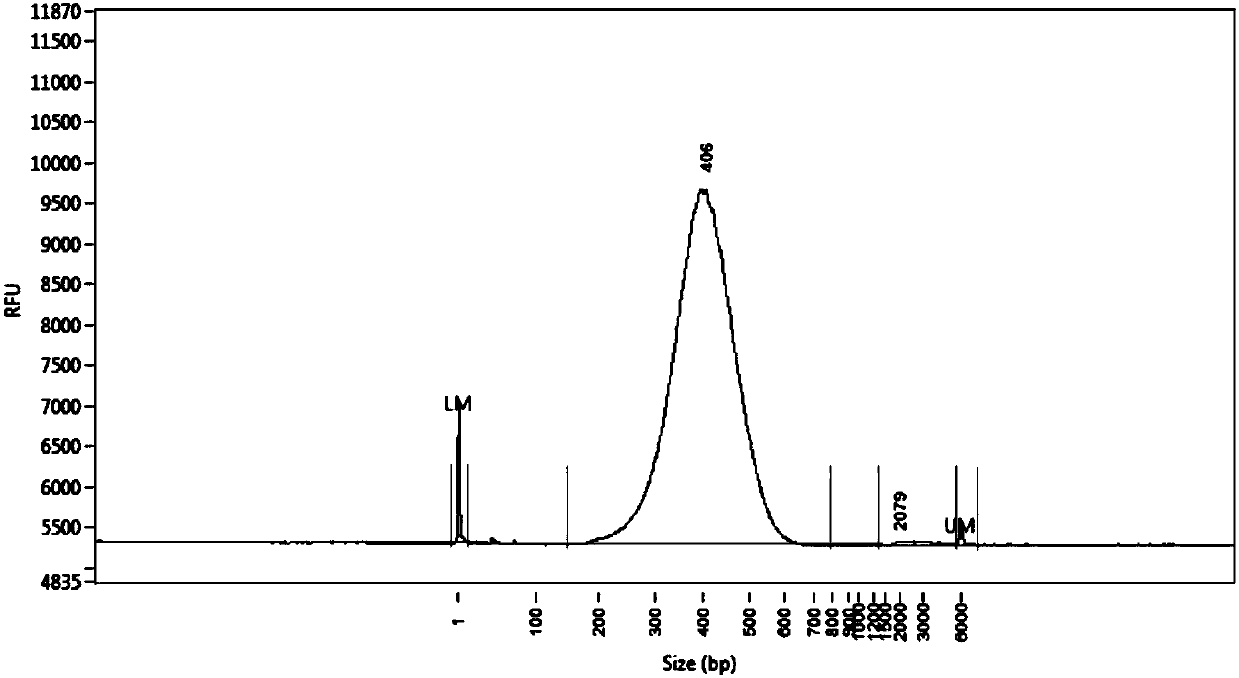
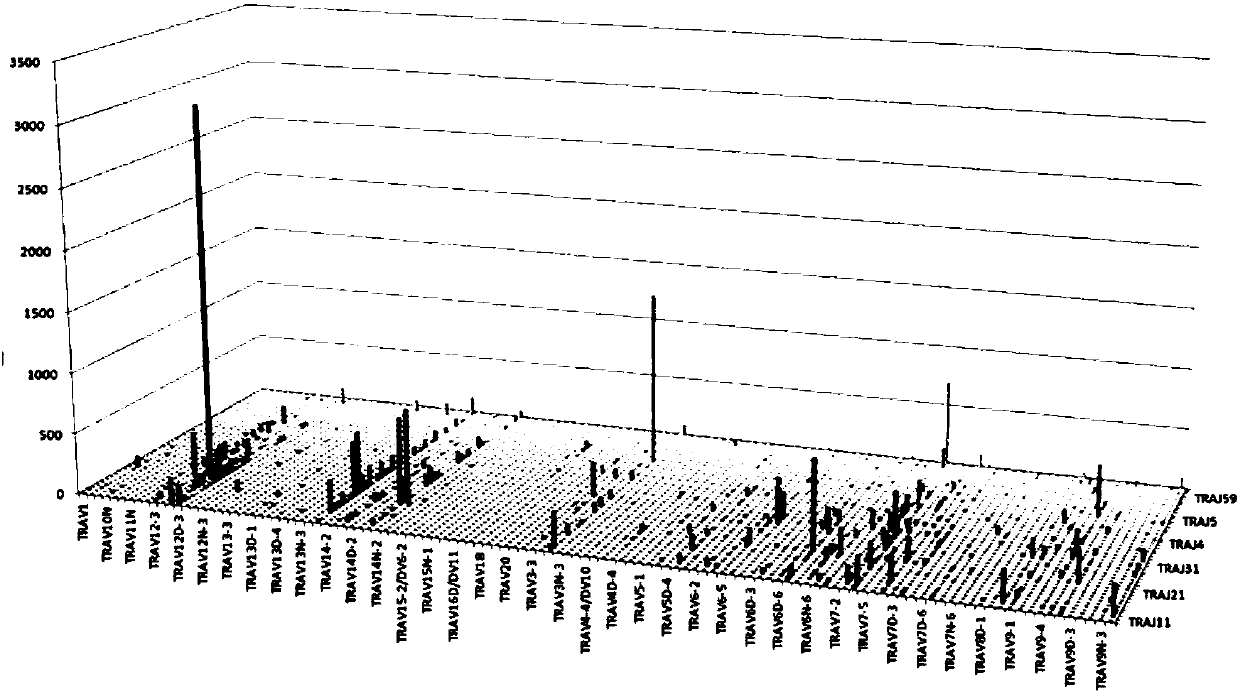
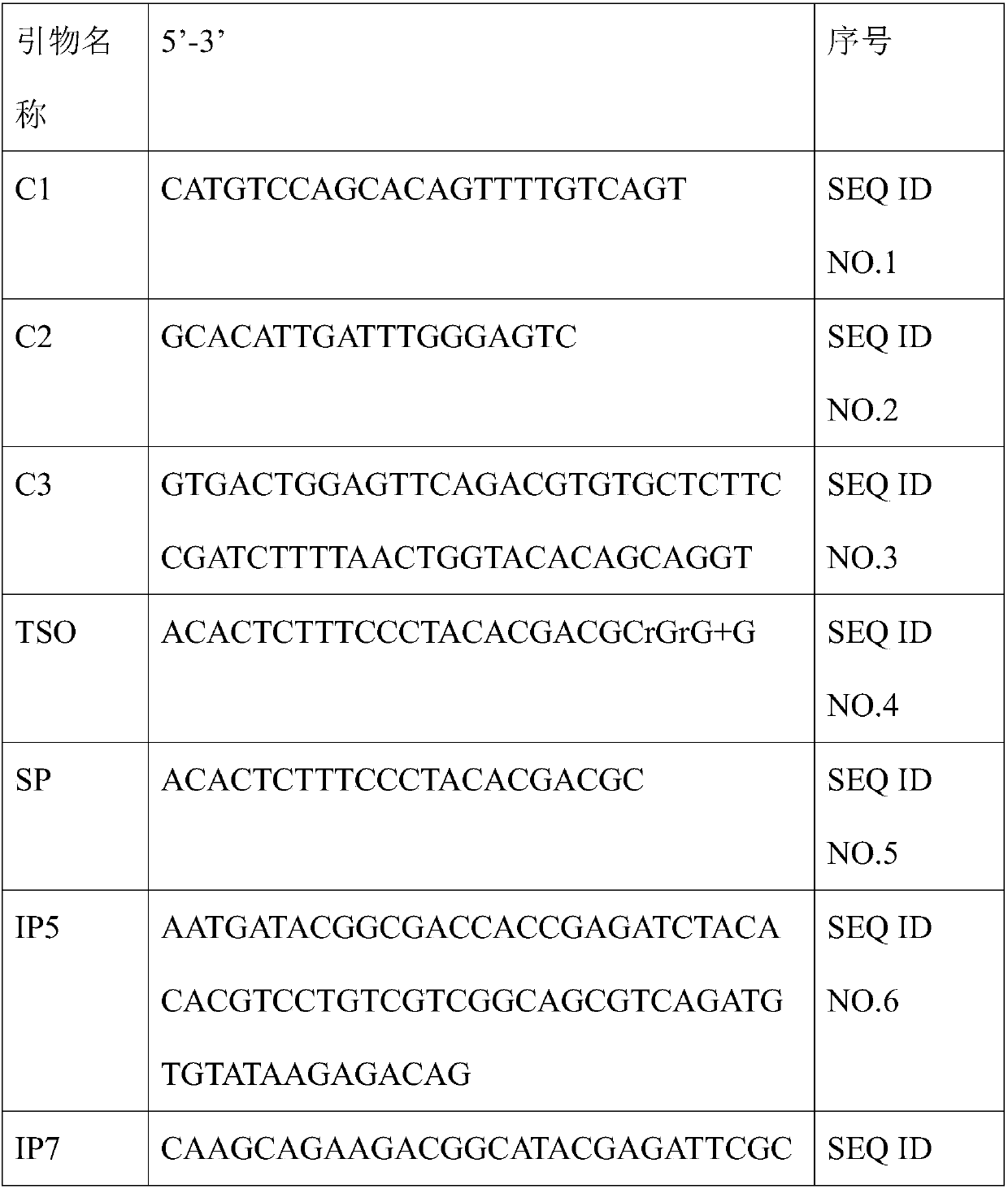
Method for constructing mouse TCR alpha CDR3 region library
A mouse and library technology, applied in the biological field, can solve problems such as product deviation, poor reproducibility, and mutual interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0023] The present invention will be described in further detail below in conjunction with specific embodiment:
[0024] The method for constructing a mouse TCR alpha CDR3 region library based on high-throughput sequencing 5' RACE combined with Tn5 transposase technology comprises the following steps:
[0025] Step 1 Extract total RNA from mouse tissue
[0026] The method for extracting total RNA in mouse tissue / whole blood using the Trizol method (Roche, Tripure Isolation Reagent) includes the following steps: Homogenize the tissue / whole blood, add Trizol, pipette, let stand at room temperature for 5min, directly extract RNA or put into- Freeze at 80°C, add 200ul chloroform / ml Trizol, invert and mix for 30s, and place at room temperature for 3min. Centrifuge at 12000g for 15min at 4°C. Aspirate the upper aqueous phase to another EP tube as much as possible, and be careful not to suck the middle layer. Add isopropanol at the rate of 0.5ml isopropanol / ml Trizol, invert and m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com