Method for performing genetic modification on DHDPR (Dihydrodipicolinic Acid Reductase) in corynebacterium glutamicum to increase lysine yield
A Corynebacterium glutamicum and genetic modification technology, applied in the fields of genetic engineering and enzyme engineering, can solve problems such as insufficient NADPH content, and achieve the effect of eliminating insufficient NADPH supply and improving capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
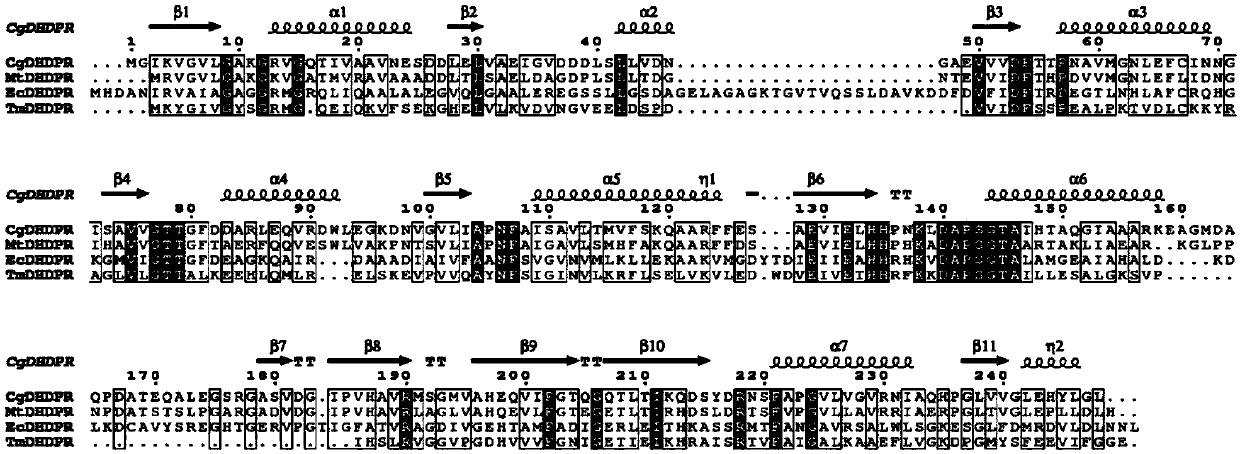
[0039] Example 1: In vitro site-directed mutagenesis of the DHDPR-encoding gene dapB in C. glutamicum
[0040] Using the C. glutamicum JL-6 genome as a template and dapB-F / dapB-R as primers, a 747bp dapB fragment was amplified by PCR reaction, and EcoRI and HindIII restriction enzymes were introduced at the 5' and 3' ends of the PCR product, respectively. cut site.
[0041] The above dapB fragment was connected to a T vector (Pucm-T), transformed into Escherichia coli, and the recombinant plasmid Pucm-T / dapB was extracted. The plasmid Pucm-T / dapB was used as a template, and the mutant primers MCB1-F / MCB1-R and MCB2-F / MCB2-R were used as primers to carry out PCR reactions, and the DNA fragment purification kit was used to purify and use restriction endonuclease DpnI Enzyme digestion (DpnI only recognizes methylated DNA, but the newly synthesized DNA is not methylated), then the enzyme digestion product is purified and transformed into E.coli JM106 competent cells, and spread o...
Embodiment 2
[0043] Example 2: Expression and purification of DHDPR mutants in E.coli BL21
[0044] According to Table 1, the recombinant plasmid Pucm-T / dapB was cut with restriction enzymes EcoRI and HindIII respectively. A31G,A32C and Pucm-T / dapB C37G,G38C . Subsequently, dapB was recovered using a gel recovery kit A31G,A32C and dapB C37G,G38C fragment. will dapB A31G,A32C and dapB C37G,G38C The fragments were respectively connected with pET28a cut with the same restriction enzyme to construct recombinant plasmid pET28a / dapB A31G,A32C and pET28a / dapB C37G,G38C ; The recombinant plasmid pET28a / dapB A31G,A32C and pET28a / dapB C37G,G38C Transfer to E.coli BL21, pick positive transformants on the kanamycin-resistant LB plate to liquid LB, collect the bacteria after IPTG induction, and after ultrasonic crushing, SDS-PAGE of the supernatant, a molecular weight of about 30kDa was detected The specific band ( figure 2 ), consistent with the reported target protein size, while the cont...
Embodiment 3
[0045] Example 3: Determination of Kinetic Parameters of DHDPR Mutants
[0046] Take the above-mentioned purified mutant DHDPR K11A 、DHDPR R13A Add wild-type DHDPR to the DHDPR enzyme activity assay reaction system, monitor the change of absorbance at 340nm in real time, and calculate the K according to the change of absorbance m and K cat (Table 2).
[0047] Reaction system: 100mmol L -1 MOPS buffer (pH 7.2), 50 μmol L -1 Dihydrodipicolic acid, 0.01~0.04mmol L -1 NADPH or 0.004~0.032mmol L -1 NADH; Reaction temperature: 30°C; Reaction time: ≥300s.
[0048] Table 2. Kinetic parameters of wild-type DHDPR and its mutants
[0049]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com