Method for detecting genes of lung cancer and colorectal cancer based on NGS method
A technology for colorectal cancer and genetic detection, applied in the direction of microbial measurement/testing, biochemical equipment and methods, etc., can solve the problems of low cost and high sensitivity, and achieve the effect of improving the possibility and solving the problem of synthesis cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The technical solution of the present invention is described in detail below. Those skilled in the art should understand that the technical solution of the invention can be modified or equivalently replaced, which should be covered by the scope of the claims of the present invention.
[0026] In this example, 15 lung cancer-related genes were selected for panel design, such as: EGFR, ALK, KRAS, BRAF, ROS1, RET, ERBB2, MET, AKT1, NRAS, PIK3CA, PTEN, DDR2, MAP2K1 (MEK1) and FGFR1, 12 genes related to colorectal cancer were also selected, such as: EGFR, KRAS, BRAF, ERBB2, MET, AKT1, NRAS, PIK3CA, PTEN, MAP2K1 (MEK1), FGFR1 and SMAD4.
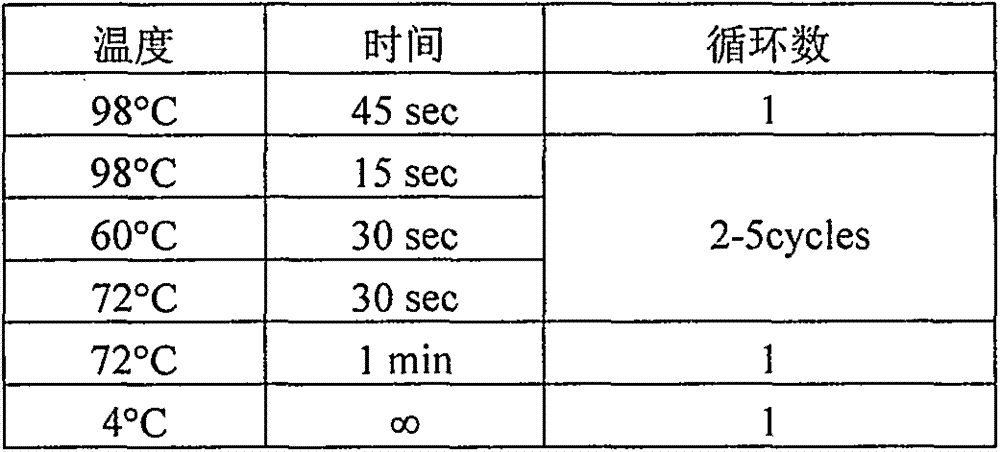
[0027] The verification method adopted in the library building hybridization capture process of the present invention is as follows:
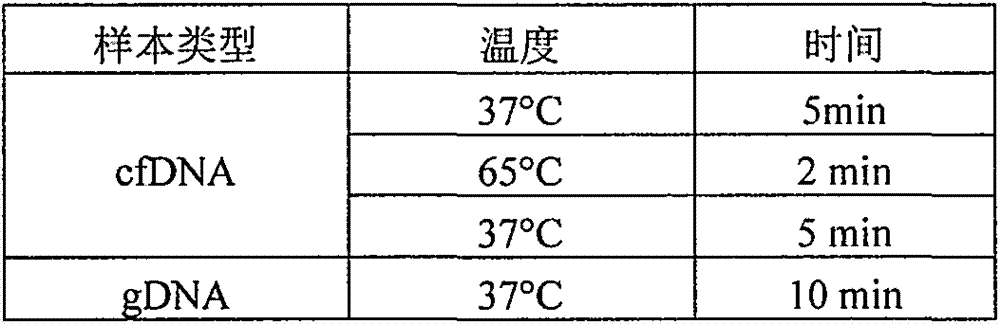
[0028] 1.1gDNA fragmentation and purification:
[0029] 1.1.1. Take 500ng according to the qubit concentration, add water to make up to 100μl, add covaris 130μl interrupted tube, set the program: 50W, 20%, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com