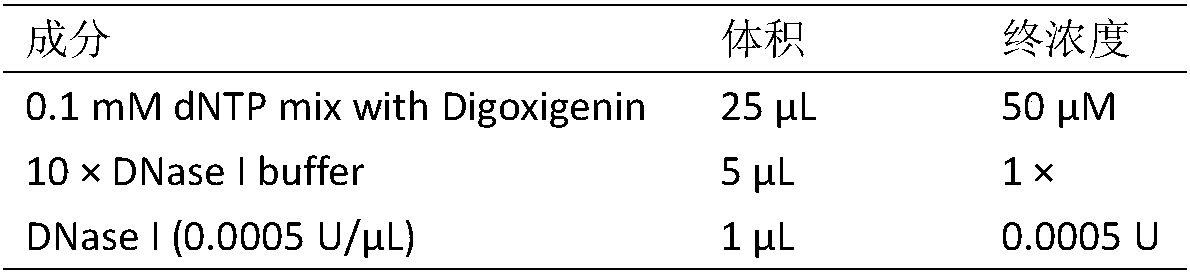Method for identifying consanguinity of saccharum arundinaceum in sugarcane via spread sequence
A sequence listing and sequence technology, which is applied in the field of plant detection and identification, can solve the problems of being unable to stably and effectively detect hybrid offspring, being unable to know and accurately detect the chromosomes or fragments of Pseudomonas chinensis, and failing to perform stability detection, etc., to achieve reliability High, high efficiency, good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Embodiment 1, a kind of method that utilizes interspersed sequence identification to comprise the blood lineage of sagegrass in sugarcane, it is characterized in that identification step is as follows:
[0016] 1. Preparation of root tip cell slides: select healthy sugarcane stems and culture them at 28°C under constant temperature and humidity. A 2 cm root tip was excised and pretreated with 1,4-dichlorobenzene for 2 h. Then transfer to Carnoy's fixative solution and fix at 4°C for 24h. The fixed apical ddH 2 O was hypotonic for 0.5 h, and at 37°C, the fixed root tips were hydrolyzed in a mixed enzyme (4% Onozuka R10cellulose, 0.5% pectolyase Y-23 and 0.5% pectinase) for 3 h. Put the root tip into Carnoy's fixative solution for 0.5h and fix it on a clean glass slide to dry to obtain the root tip slide.
[0017] 2. Preparation of labeled probes: The reaction system for Digoxigenin labeled interspersed sequences is shown in Table 1:
[0018] Table 1 Reaction system o...
Embodiment 2
[0027] Example 2, using the PCR method to mutually confirm the interspersed sequence to detect the ancestry of Banmao
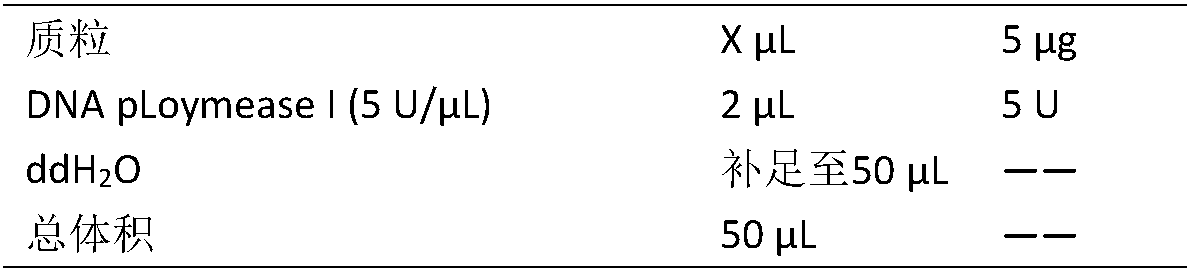
[0028] In order to further simplify the detection process, we designed and screened a pair of amplification primers 128F and 128R with reference to the specific scattered sequence of P. chinensis, the nucleotide sequences of which are shown in SEQ ID NO: 2 and SEQ ID NO: 3 in the sequence list Nucleotide sequence; 70 parts of plant material genomic DNA were extracted by CTAB method, including 5 parts of the original species of Banmao, tropical species of Saccharum, wild species with large stems, wild species with thin stems, Chinese species, Indian species and cultivated varieties of sugarcane ( As shown in Table 3), the hybrid progeny of Banmao and sugarcane F 1 、BC 1 、BC 2 、BC 3 A total of 35 materials (as shown in Table 4), OD 260 / OD 280 The quality of the DNA whose ratio is between 1.6 and 1.8 meets the requirements of the experiment, and the concen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com