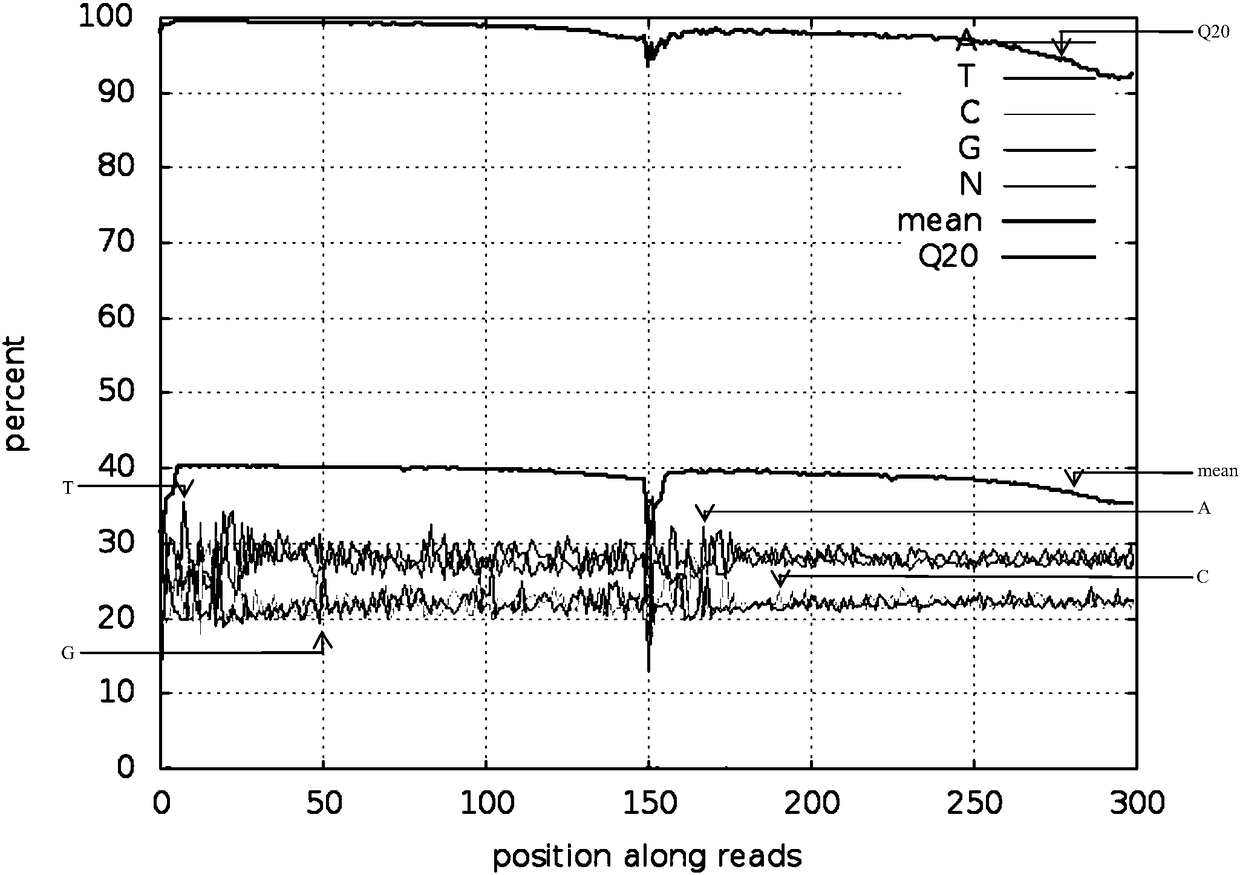
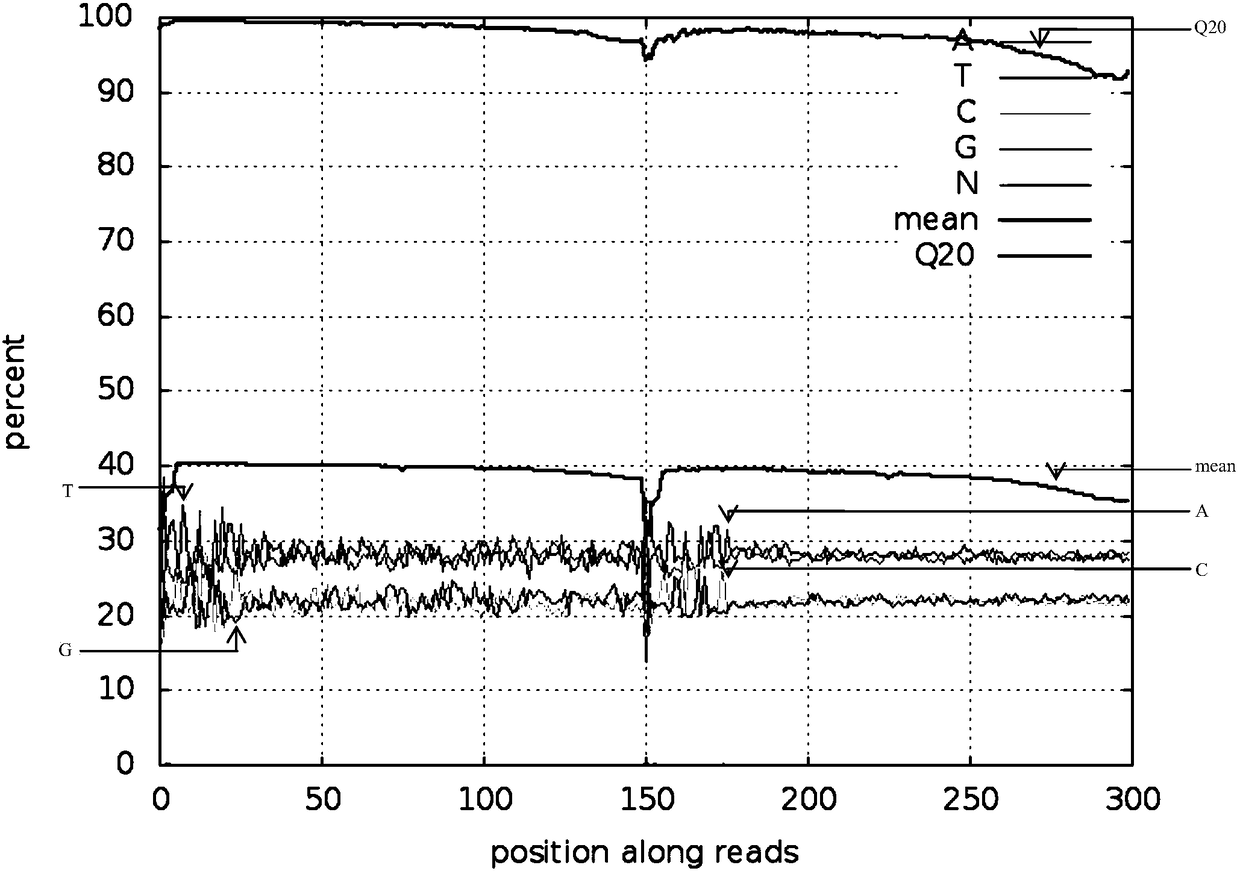
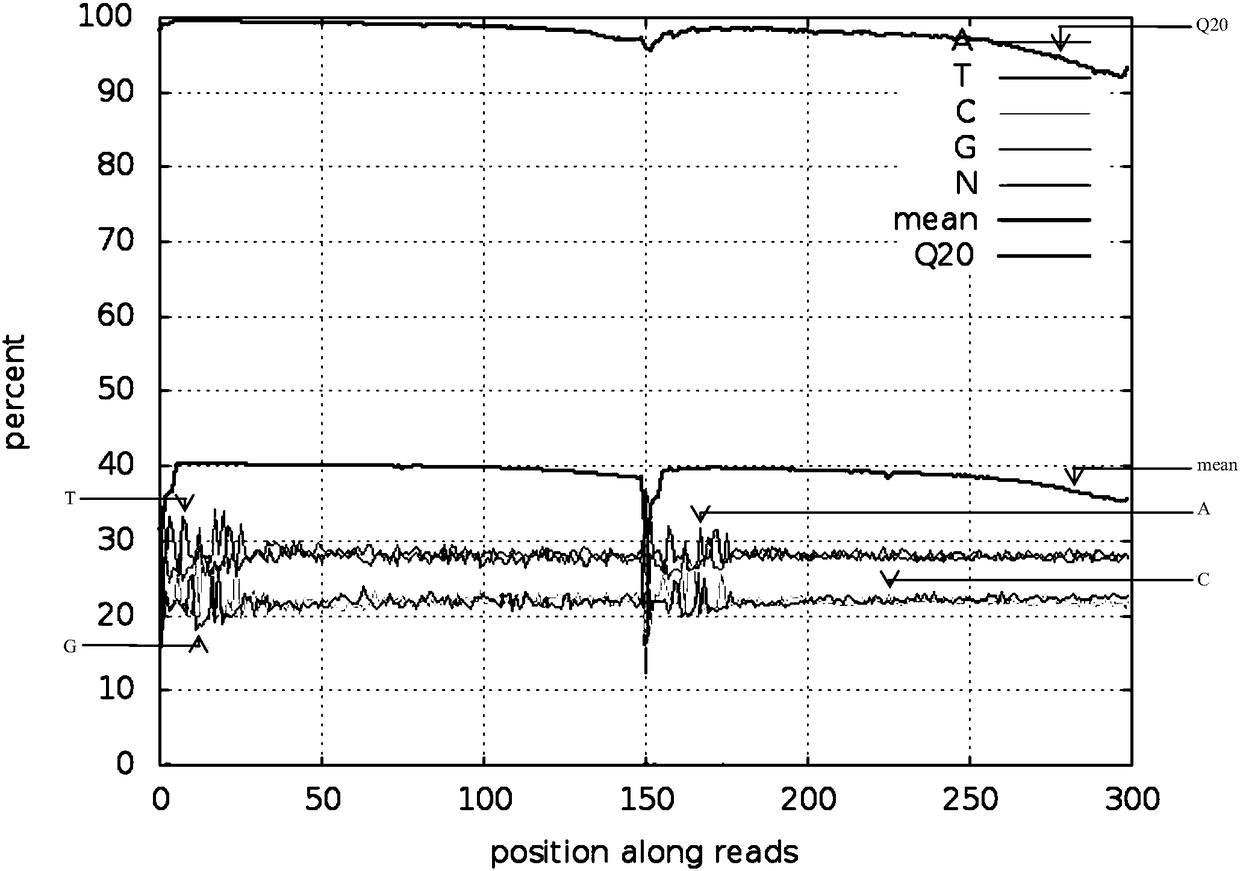
Reduced genome library building method for millet
A millet and gene technology, applied in the field of simplified gene library construction of millet, can solve the problems of unstable sequencing quality, unsatisfactory, low throughput, etc., and achieve the effect of improving data utilization and avoiding two-way complementarity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 is a simplified gene library method for 6 millet samples,
[0059] Selection of materials: select 6 millet samples, extract the genomic DNA of the millet samples, and the extraction method is carried out according to the operation manual of the new plant extraction kit of Tiangen Biochemical Technology Co., Ltd.;
[0060] Restriction endonuclease ApeK Ⅰ: Biolab company;
[0061] NEB Buffer (NEB Buffer): Takara Company;
[0062] T4 DNA ligase (T4 DNAigase): Takara company;
[0063] 10xT4 DNAigase Reaction Buffer: Takara Company;
[0064] HiFi enzyme: Kakp company;
[0065] AMPure XP Beads: Novozyme Biotechnology Co., Ltd.
[0066] The simplified gene library method of 6 millet samples specifically includes the following steps:
[0067] Step 1. Using the restriction endonuclease ApeK Ⅰ to digest the genomic DNA of several millet samples respectively to obtain digested products;
[0068] The enzyme digestion system used is: NEB Buffer (NEB Buffer) 2μL, A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com