EST-SSR molecular marker developed based on lycoris aurea transcriptome sequence and application thereof
A technology of transcriptome sequence and labeling, which can be used in recombinant DNA technology, DNA/RNA fragments, determination/inspection of microorganisms, etc., and can solve problems such as non-specific molecular markers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
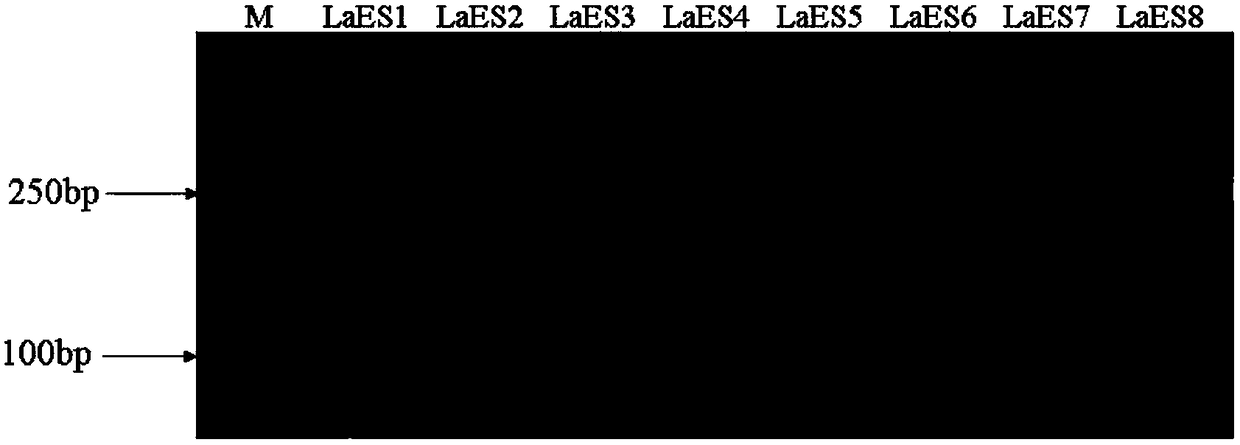
[0021] Example 1 : 20 SSR markers developed from the Smiley Transcriptome were obtained by the following method:
[0022] (1) Obtain the EST sequence according to the transcriptome sequencing results of Hudi Xiao, use the software Microsatellite (MISA) to find the SSR sequence and design primers. The principle of primer design is that the length of the PCR product is 100-350bp; the GC content is 40-70%, and the maximum Suitable for 50%; annealing temperature 50-65 ℃; primer length: 19-22bp, the primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd.;
[0023] (2) Harvest the young leaves of Hudixiao, and the information on the germplasm resources of Hudixiao is shown in Table 2. The DNA of the sample to be tested was extracted by the Biotech Plant Genomic DNA Rapid Extraction Kit, and the operation was carried out with reference to the instructions of the kit. Dilute it by 50 times, check the DNA concentration by spectrophotometer, check the quality by electrophor...
example 2
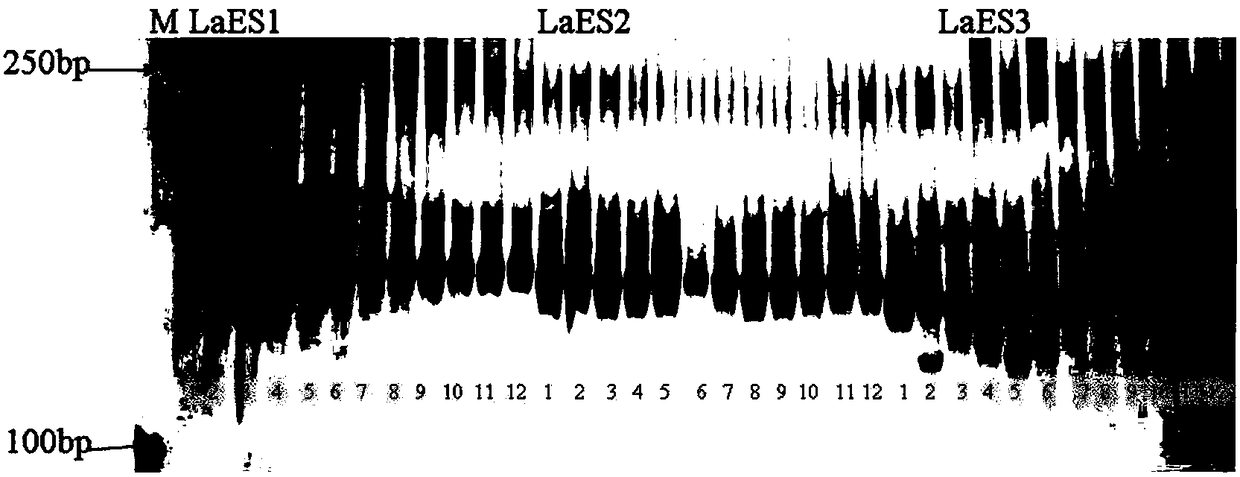
[0037] Example 2: Genetic diversity analysis of 20 EST-SSR markers applied to the material in Table 2
[0038] (1) Extract the genomic DNA of the material in Table 2, and adjust the DNA concentration to 10ng / μL;
[0039] (2) Using the above DNA as a template, use the primers shown in SEQ ID NO. 1 to 40 in Sequence Table 1 to carry out PCR amplification, and add 2.0 μL of 10x Taq Buffer (containing Mg to the 20 μL reaction system respectively). 2+ ), 1.6μ of 2.5nM dNTP, 5units / μL Taq DNA polymerase 0.1μL, forward primer 4pmol, reverse primer 4pmol, 10ng / μL DNA template 1μL, ddH 2 O supplement the system to 20 μL. DNA amplification was performed using an Eppendorf Mastercycler. The specific reaction procedure was: pre-denaturation at 94°C for 3 min, followed by 35 cycles of 94°C (30S) / 58°C (30s) / 72°C (30S), and a final extension at 72°C for 10 min;
[0040] (3) The PCR amplification product in step (2) was detected by 8% polyacrylamide gel electrophoresis, the voltage was 180V...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com