Microbial production of nicotinamide riboside
一种烟酰胺、核糖苷的技术,应用在烟酰胺核糖苷的微生物生产领域
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
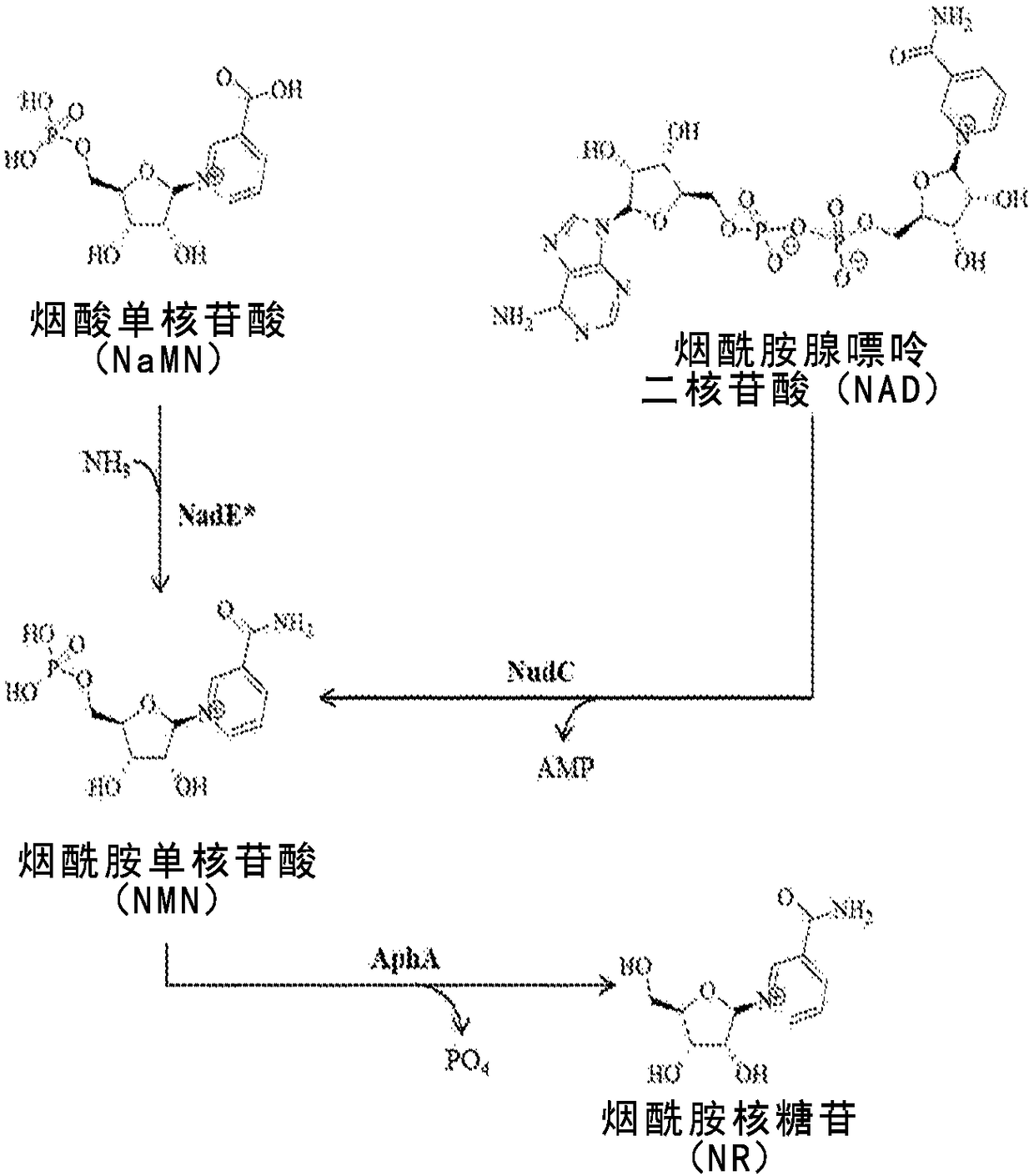
[0224] Identification of sequences encoding NaMN amidation activity (NadE*)
[0225] Sorci and collaborators identified the enzyme FtNadE* (SEQ ID NO: 1) encoded by the genome of Francisella tularensis and demonstrated its ability to function as a nicotinamide mononucleotide (NaMN) amidase in vivo and in vitro (Sorci L.e., 2009). Furthermore, they propose that three amino acid residues are responsible for the enzyme's substrate preference for NaMN (vs. NaAD): Y27; Q133 and R236. To identify additional sequences encoding this function, 50 unique sequences obtained from a BLAST search of the NCBI nr / nt database on September 14, 2016 using the amino acid sequence of FtNadE (SEQ ID NO: 2) using the default parameters of tBlastn will be used. Nucleotide sequences were translated and aligned using the Geneious alignment algorithm (Biomatters, LLLC.). Sixteen of these sequences have conserved tyrosine, glutamine, and arginine (i.e., contain a "Y-Q-R motif") that align with Y27, Q...
Embodiment 2
[0227] Gene construct for expressing NaMN amidation activity (NadE*) in E.coli
[0228] Based on maximizing the phylogenetic distance ( Figure 6 ), select 10 sequences encoding predicted NadE* open reading frames (SEQ ID NO: 2 and 19 to 27) in 16 predicted NadE* gene sets, and perform codon optimization for expression in E.coli, The Geneious codon optimization algorithm was used, using the E.coli K-12 codon usage table, and the rare threshold was set to 0.4. The optimized sequences (SEQ ID NOs 28 to 37) were synthesized de novo by GenScript, Inc. and cloned by GenScript likewise into XhoI / NdeI digested pET24a(+) (Novagen, Inc.), resulting in the plasmids in Table 1 . The plasmid was transformed into BL21(DE3), which allowed IPTG induction of the NadE* gene to induce NR synthesis and resulted in strains ME407, ME644, ME645, ME646, ME647, ME648, ME649, ME650, ME651, ME652 (Table 2).
[0229] Table 1: Plasmids used in this study
[0230]
[0231]
[0232] Table 9: ...
Embodiment 3
[0240] Characterization of E. coli strains expressing NadE* enzyme
[0241]To examine the effect of NadE* expression on NR production, E. coli strains were inoculated from single colonies into LB medium and grown overnight (2 mL, 37 °C, 15 mL tube, 250 rpm, 50 ug / mL kanamycin). The preculture (200 μL) was used to inoculate 2 mL of M9nC medium with or without 25 μM IPTG and grown in 24-well deep-well plates (Whatman Uniplate, 10 mL, round bottom) sealed with AirPore tape sheets (Qiagen) for 3 days (Infors Multitron shaker, 800 rpm, 80% humidity). Samples were analyzed by LC-MS as described herein. In the absence of plasmid, NR production was below the limit of quantitation in the presence and absence of induction with 25 [mu]M IPTG. After induction, strains containing the plasmid for expression of the NadE* enzyme produced up to 2.7 mg / L of NR (Table 3).
[0242] Table 3: Nicotinamide riboside concentrations (mg / L) in E. coli shake plate cultures after IPTG induction (aver...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com