Cancer marker detection method combining DNA coding technology with nanopore technology
A DNA molecule and antibody detection technology, applied in recombinant DNA technology, biochemical equipment and methods, DNA/RNA fragments, etc., can solve problems such as inapplicability of antigens and coding errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
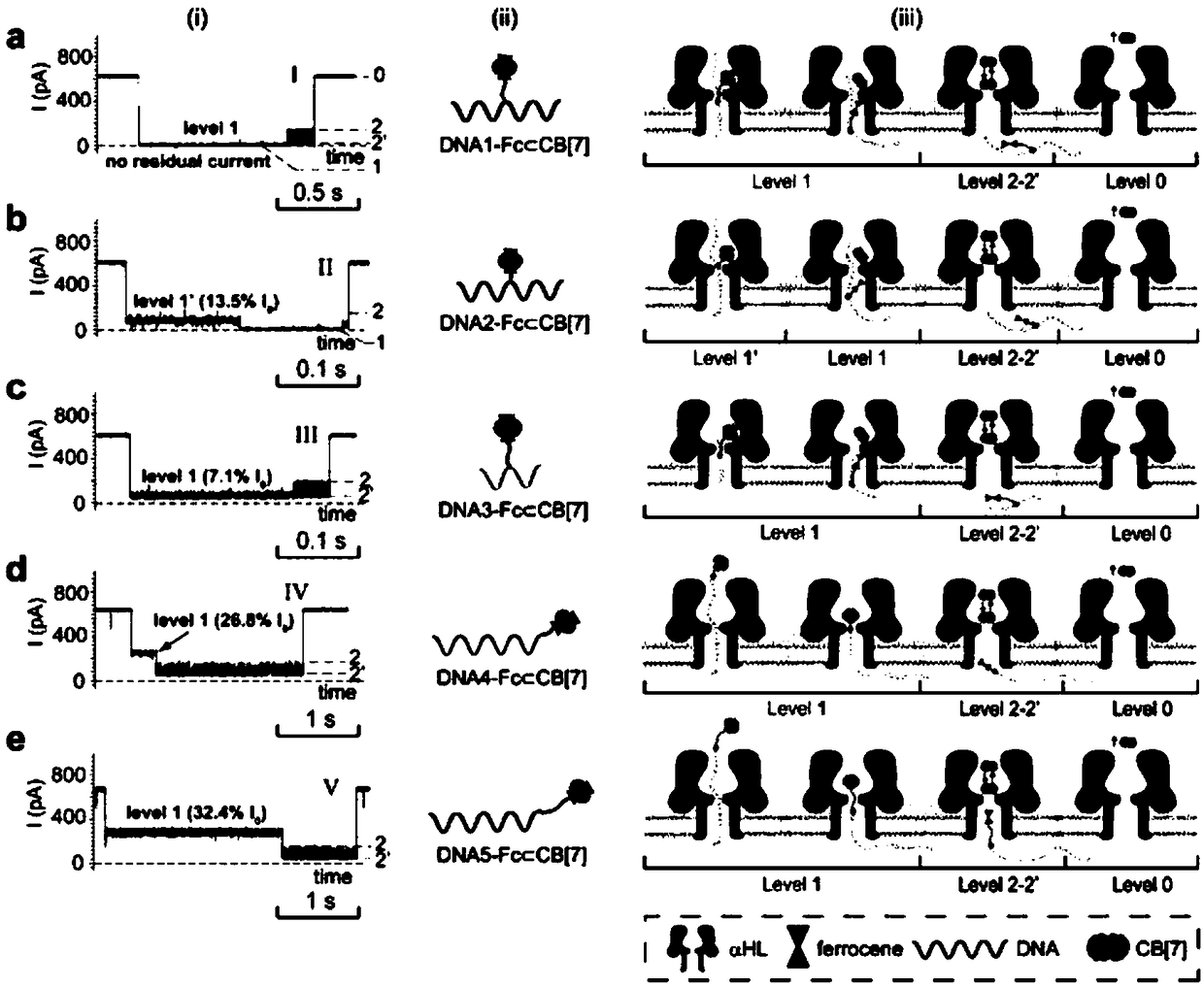
[0140] Embodiment 1, the synthesis of five kinds of probe molecules
[0141] 1. Artificially synthesize the five single-stranded DNA molecules listed in Table 1.
[0142] Table 1 Five coding DNA sequences and modification methods
[0143]
[0144] 2. Perform the following operations on the five single-stranded DNA molecules synthesized in step 1:
[0145](1) Take 3.3 μL DNA molecule solution (solvent is water, the concentration of single-stranded DNA molecule prepared in step 1 is 100 μM), 1.2 μL deionized water, 2 μL azide ferrocene solution (solvent is acetonitrile, azide The concentration of iron is 200mM), 1 μL sodium ascorbate solution (the solvent is water, the concentration of sodium ascorbate is 20mM), 0.5 μL copper nitrate solution (the solvent is water, the concentration of copper nitrate is 20mM), 2 μL HEPES buffer solution (200mM) mixed , a total of 10 μL of the mixed solution.
[0146] (2) The mixed solution obtained in step (1) was vortex reacted at room te...
Embodiment 2
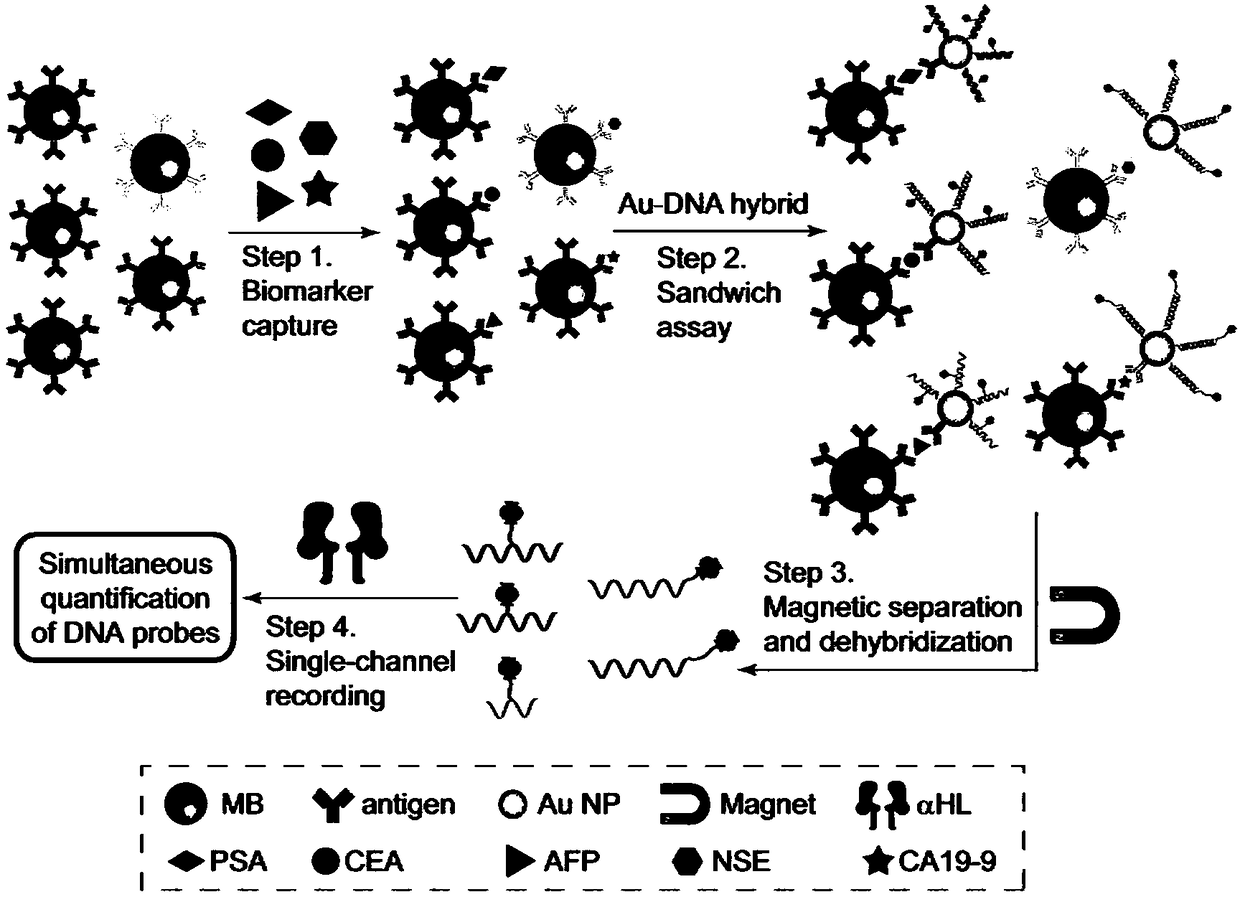
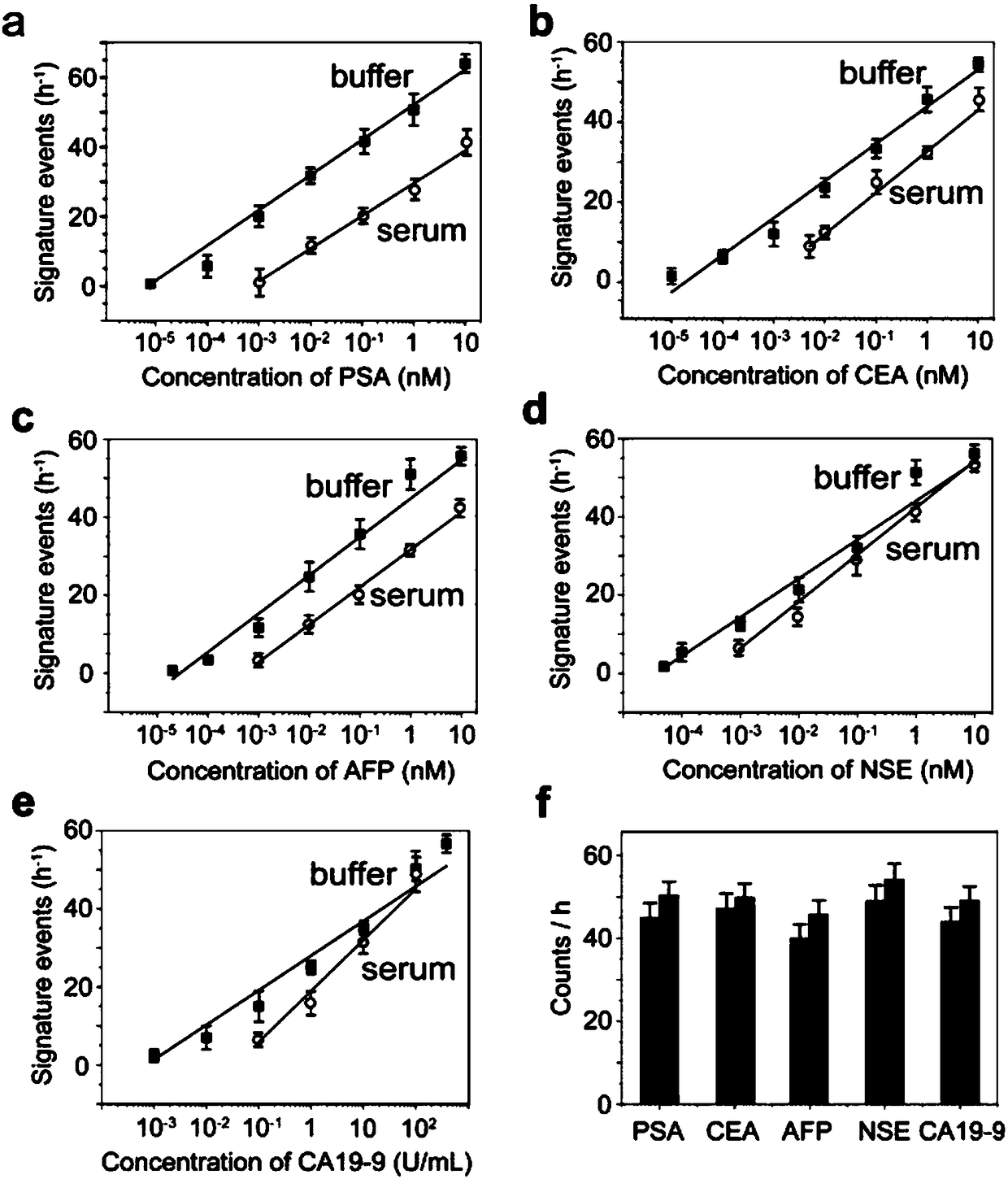
[0160] Embodiment 2, establishment of detection method
[0161] The principle of simultaneous detection of five antigens by using the above five probes is shown in figure 2 . The antigen to be tested can be any antigen molecule.
[0162] The following experiments use five cancer markers PSA (prostate specific antigen), AFP (alpha-fetoprotein), CEA (carcinoembryonic antigen), NSE (neuron-specific enolase) and CA19-9 (cancer antigen CA19-9 ) as an example.
[0163] 1. Detection of antibody-modified gold nanoparticles
[0164] 1. Take 1ml colloidal gold solution (particle size 30nm, concentration 2×10 11 particles / mL), with 0.1M NaHCO 3 Adjust the pH of the colloidal gold solution to 9.2 with the aqueous solution, add a detection antibody corresponding to the antigen to be tested into the solution, and place it at 10°C and 90 r / min for 30 minutes to obtain a colloidal gold solution modified with the detection antibody.
[0165] The concentrations of different antibodies in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com