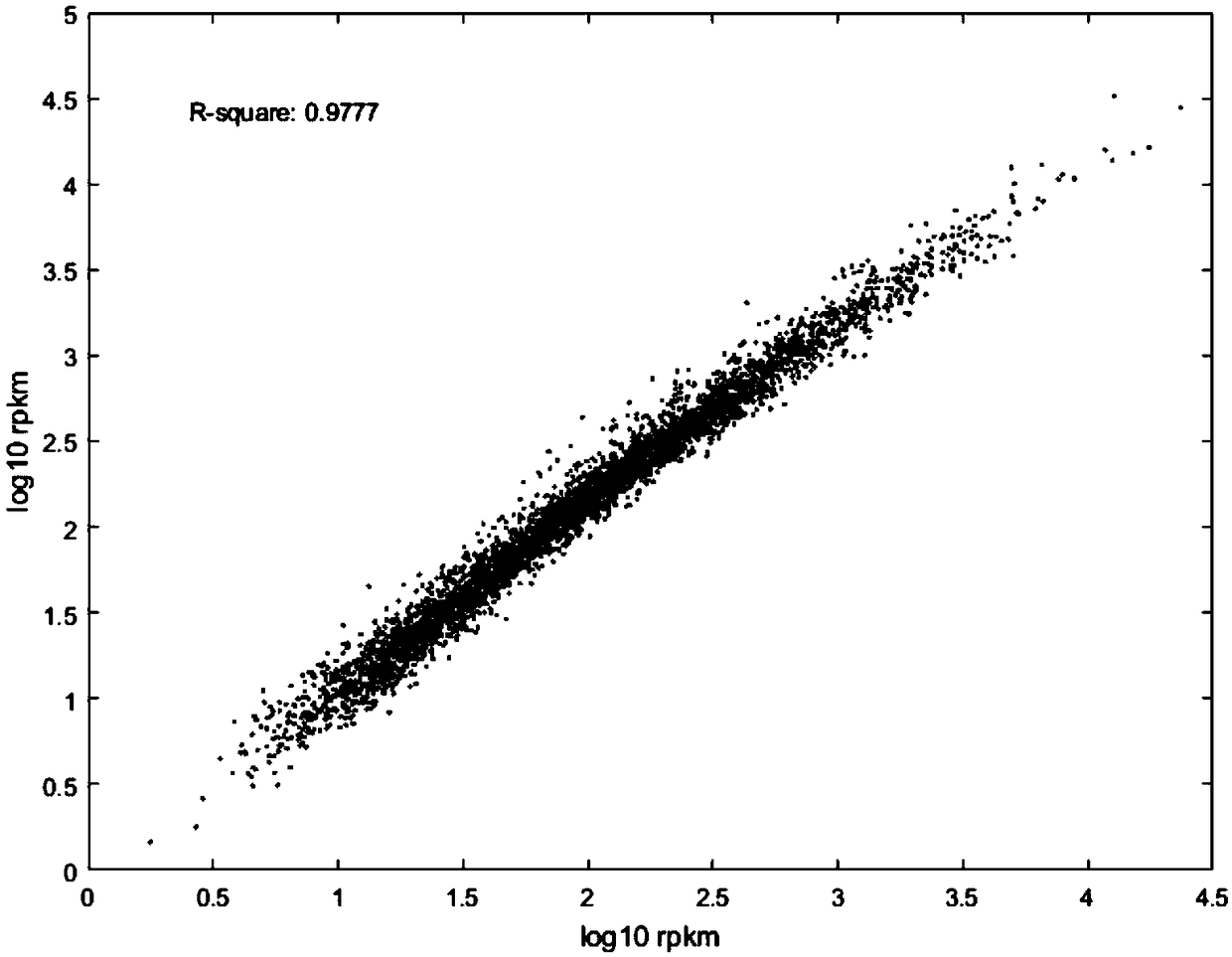
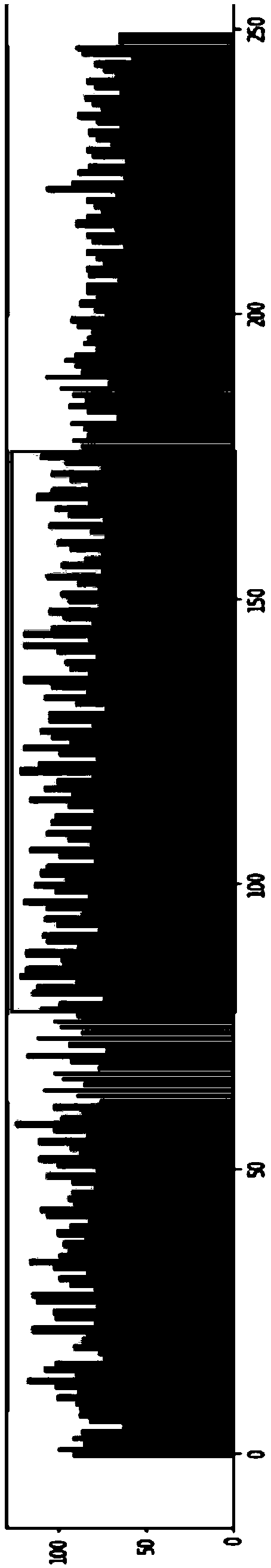
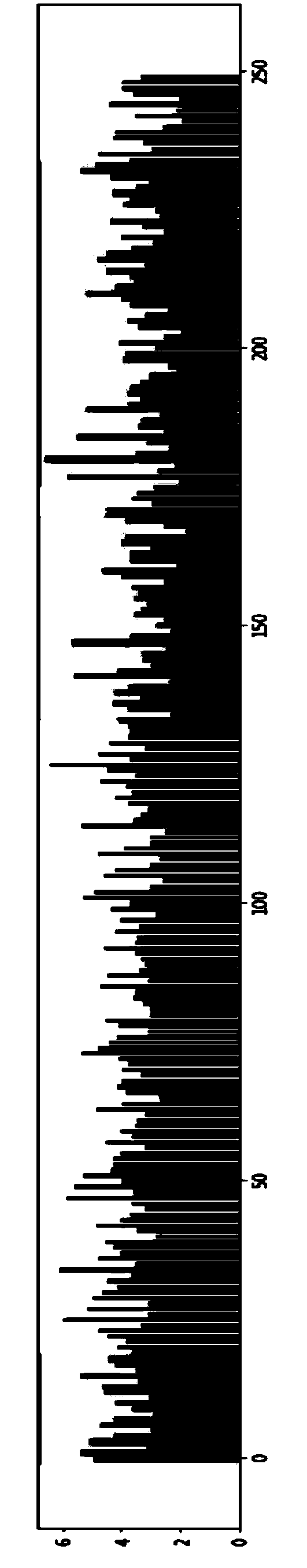
Method for constructing Ribo-seq sequencing libraries
A ribo-seq and sequencing library technology, applied in chemical libraries, biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as large applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] (1) Escherichia coli BW25113 glycerol species was streaked on the LB plate and cultivated overnight;
[0088] (2) Pick a single colony on the plate, inoculate in 3ml LB, and activate overnight at 37°C and 200rpm;
[0089] (3) Inoculate the activated seed liquid with an inoculum amount of 1%, inoculate it in 50ml of LB, and cultivate it at 37°C and 200rpm until the culture medium OD 600 =0.6;
[0090] (4) Add chloramphenicol with a final concentration of 100 μg / ml to the culture medium obtained in the upward step, shake it in ice water for 5 minutes to rapidly cool to 4°C, centrifuge at 5000g for 5 minutes at 4°C, and remove the supernatant;
[0091] (5) Add pre-cooled 50ml PBS (100μg / ml chloramphenicol) to the precipitate obtained in the upward step, resuspend the precipitate, centrifuge at 5000g at 4°C for 5min to collect the bacteria, and discard the supernatant;
[0092] (6) Repeat step (5);
[0093] (7) Add pre-cooled 5.4ml lysozyme (lysozyme) buffer to the preci...
Embodiment 2
[0124] (1) Human lung cancer A549 cells (1 million) frozen at -80°C were thawed in ice water for 30 minutes;
[0125] (2) Add pre-cooled PBS to resuspend the A549 cells obtained in the previous step, and centrifuge at 4000g for 5min at 4°C;
[0126] (3) Add 2ml of lysis Buffer to the cell pellet obtained in the previous step, let stand on ice for 30min, and centrifuge at 16800rpm for 15min at 4°C;
[0127] (4) with embodiment 1 step (9);
[0128] (5) Slowly absorb the supernatant with a pipette gun, add 0.2ml of pre-cooled M buffer Mix (50mM Tris-HCl (pH=7.9), 5mM CaCl 2 , 6mM MgCl 2 , 100μg / ml cycloheximide, 1X BSA) to wash the tube wall other than the transparent precipitate, discard it, then add 0.25ml of pre-cooled M buffer Mix to gently blow and resuspend the transparent precipitate, transfer it to an EP tube, and obtain RNC heavy suspension;
[0129] Steps (6)-(10) are respectively successively with embodiment 1 steps (11)-(15);
[0130] (11) In order to check the r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com