circRNA_26852 and application thereof
A technology of reagents and pigskin, applied in the field of circRNA_26852 and its application in pig molecular assisted breeding, can solve unclear problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1c
[0042] Example 1 circRNA sequencing and selection of differentially expressed circRNAs
[0043] 1.1 Experimental animals:
[0044]In this experiment, large white pigs and Laiwu pigs with obvious differences in fat deposition were used as materials. They were raised in Laiwu Daqian Agriculture and Animal Husbandry Co., Ltd., grown and fattened under the same feeding conditions and environment, and fed according to the nutritional requirements standard (National Research Council, NRC, 1998) To feed the diet, choose 3 large white pigs (about 100kg) and 3 Laiwu pigs (about 35kg) each with a similar weight in the species, which are 180 days old, healthy and in good condition.
[0045] 1.2 Sample collection:
[0046] After slaughtering the experimental pigs, the subcutaneous adipose tissue was quickly separated. To reduce RNA degradation, all processes were performed on ice. Afterwards, the tissue was cut into small pieces with sterile scissors, put into 5mL cryopreservation tube...
Embodiment 2
[0071] Example 2 In-depth analysis of circRNA
[0072] 2.1 Functional analysis of differentially expressed circRNA:
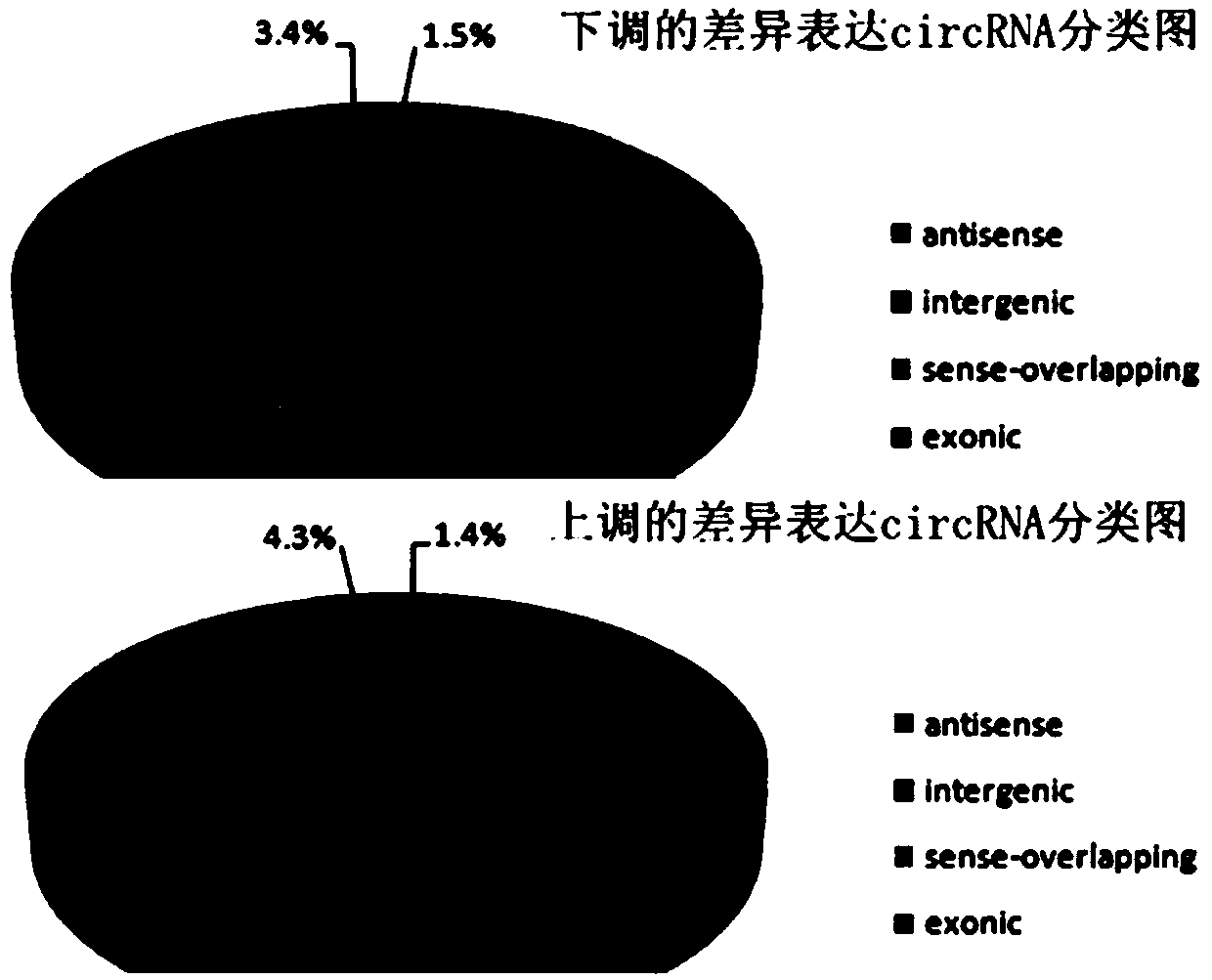
[0073] After obtaining the differentially expressed circRNAs, we used the information of the circRNA source genes to perform GO enrichment analysis on the differentially expressed circRNAs. The number of differential circRNAs included in each GO entry was counted, and the significance of differential circRNA enrichment in each GO entry was calculated using the hypergeometric distribution test method. Using the KEGG database to perform Pathway analysis of differential circRNA source genes, Pathway entries enriched for differential circRNA source genes can be found, and potential differential circRNAs in different samples may be related to changes in cellular pathways.
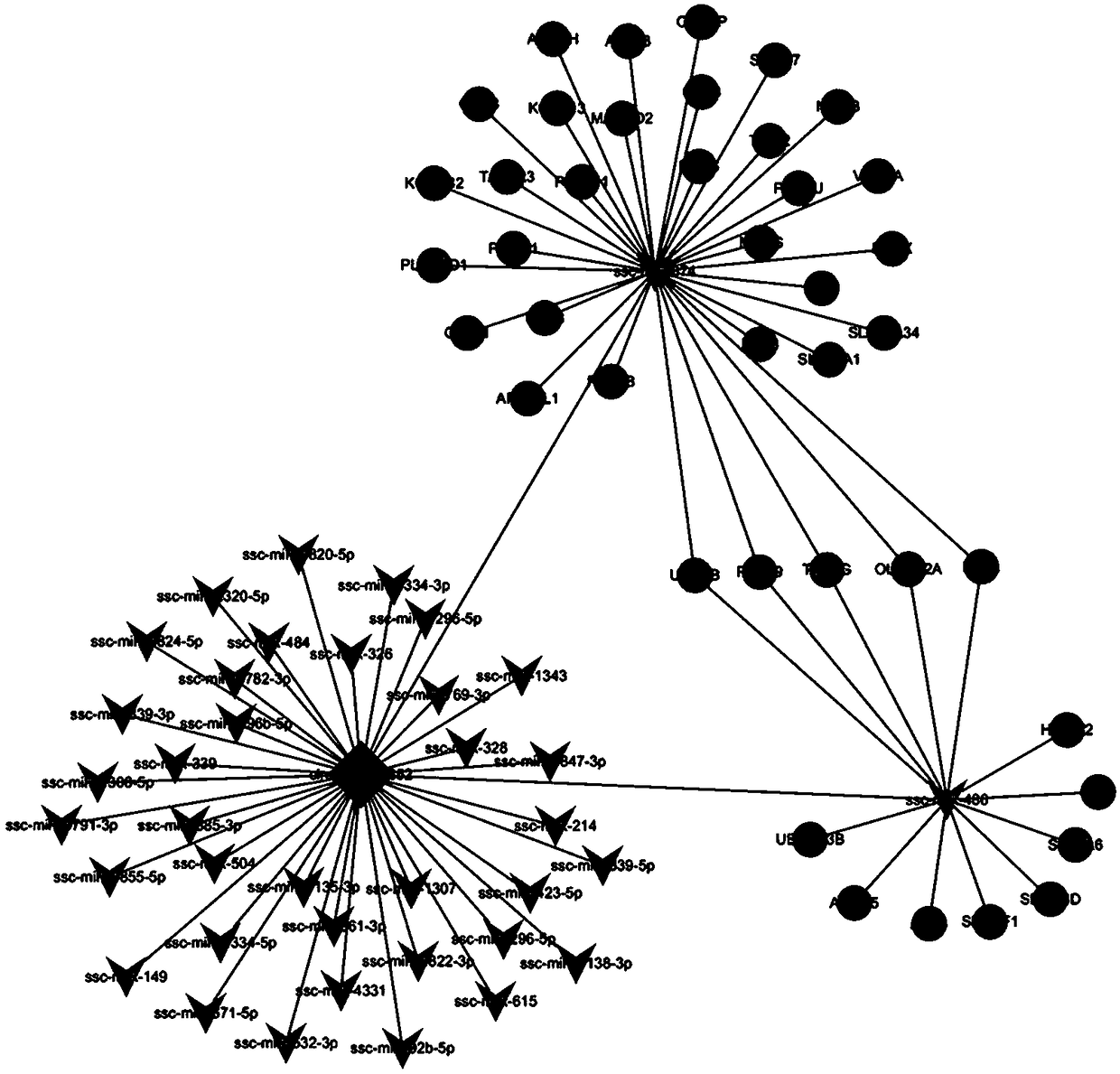
[0074] 2.2 circRNA-miRNA-mRNA interaction network diagram
[0075] The circRNA_26852 down-regulated in Laiwu subcutaneous adipose tissue was selected for subsequent analysis, combined with bioi...
Embodiment 3
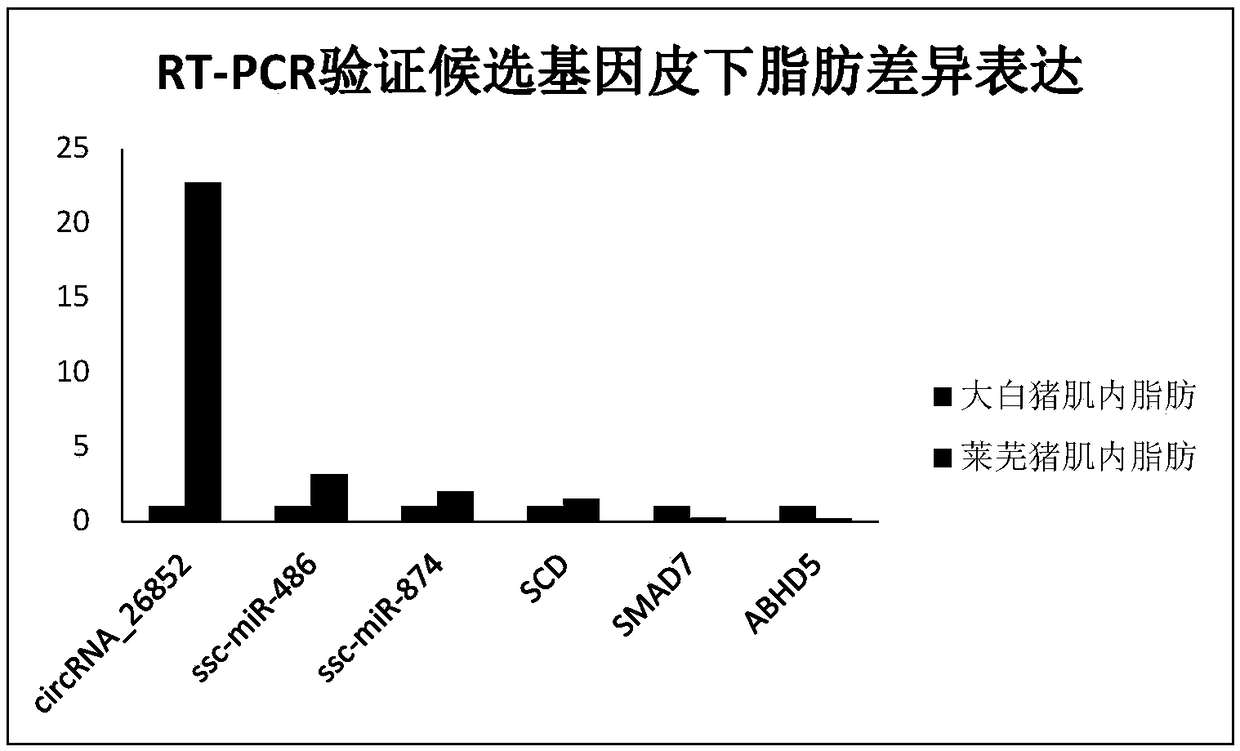
[0078] Embodiment 3 real-time fluorescent quantitative PCR verification
[0079]Select circRNA_26852, 2 miRNAs (ssc-miR-486, ssc-miR-874) in the target gene of circRNA_26852 and related genes SCD, SMAD7, ABHD5, each setting 20 biological replicates (Large White pig and Laiwu pig each 20 Example), using glyceraldehyde-3-phosphate dehydrogenase (glyceraldehyde-3-phosphate dehydrogenase, GAPDH) gene and 5S rRNA as internal references, respectively, qRT-PCR method was used to verify the expression level of the gene.
[0080] 3.1 Primer design:
[0081] circRNA_26852 amplification:
[0082] Forward primer: 5'-CACCCTGTGAGAGCCAGATTTA-3' (SEQ ID NO.2)
[0083] Reverse primer: 5'-CTGAAGCGCTGTAGTACCTGG-3' (SEQ ID NO.3)
[0084] SCD amplification:
[0085] Forward primer: 5'-CCTTTATTCTTCTCTAACCCGT-3' (SEQ ID NO.4)
[0086] Reverse primer: 5'-TCAGTGAGCAGAGAGACTTGTG-3' (SEQ ID NO.5)
[0087] ABHD5 amplification:
[0088] Forward primer: 5'-GAGGAGATGGACTCCACG-3' (SEQ ID NO.6)
[008...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com