Non-competitive probe design method, detection method and application applied to SNP typing
A detection method, a non-detection technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of detection failure, increase the intensity of detection work, and require high probe specificity, so as to reduce the workload Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
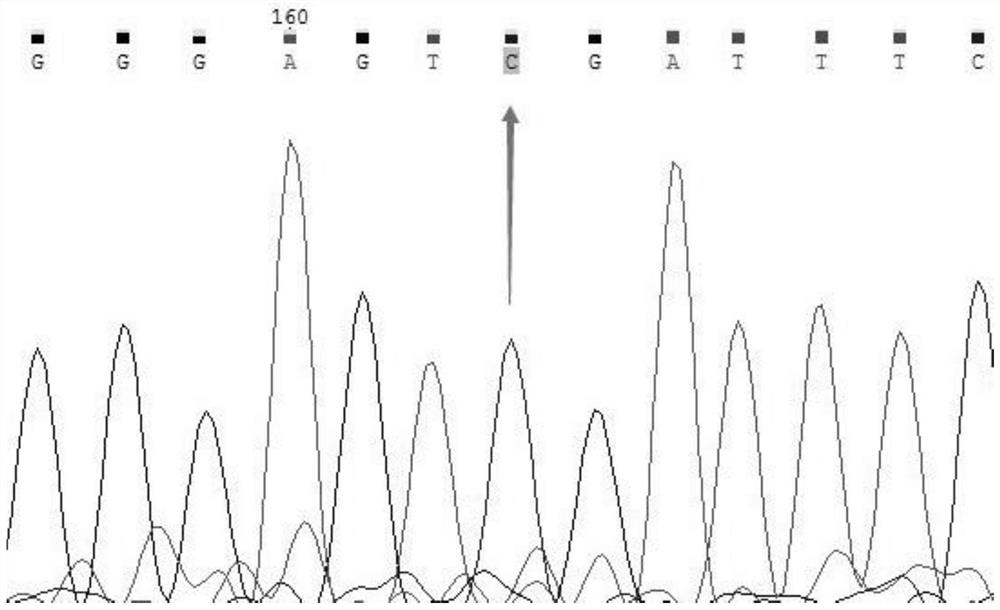
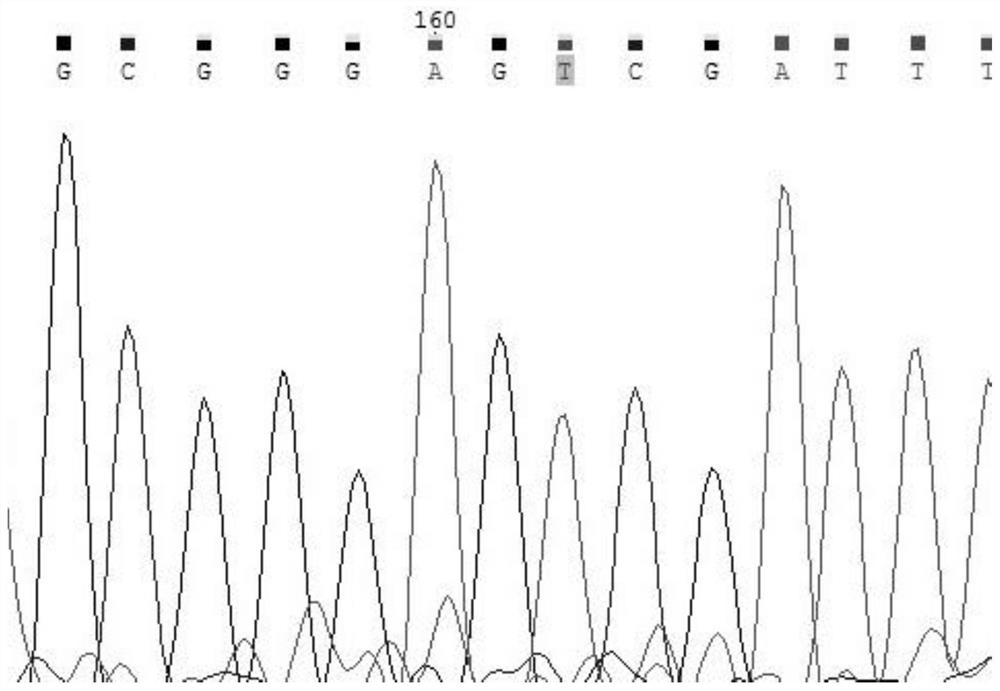
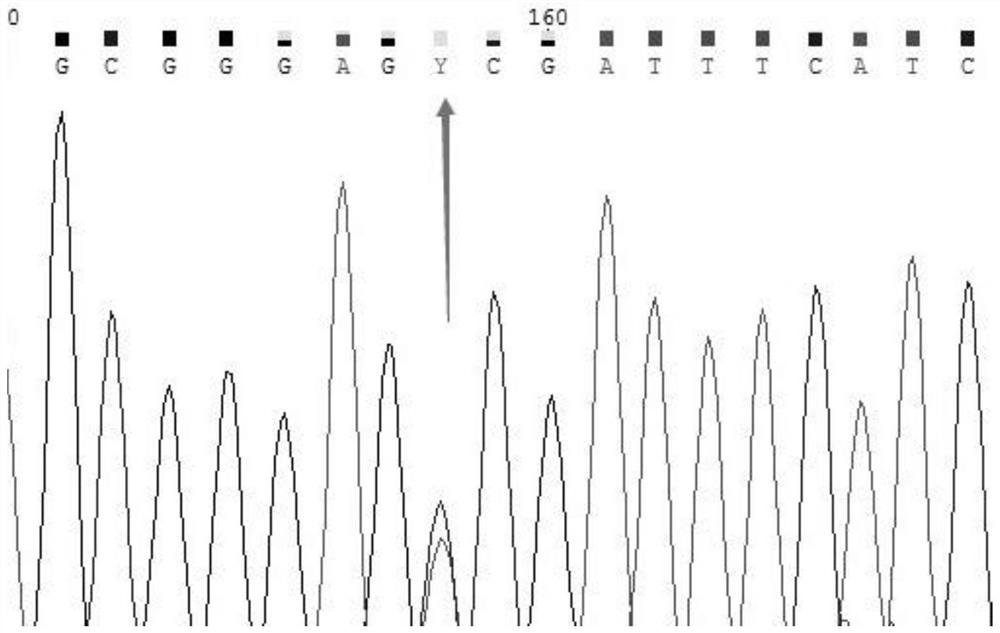
[0040] Take the detection of MTHFR gene 677 as an example. MTHFR (methylenetetrahydrofolate reductase) is one of the key enzymes of homocysteine. There are three types of genotypes at 677: wild-type CC / hybrid Synzygous mutant CT / homozygous mutant TT.
[0041]The action of MTHFR metabolizes and removes homocysteine, a toxic amino acid that damages the cell wall of the endothelium. The clinical significance of detecting the MTHFR gene 677 site is as follows: look for possible hereditary thrombosis tendency, supplement folic acid, vitamin B6 and B12, and avoid recurrent miscarriage and thrombosis.
[0042] The selected samples come from: oral exfoliated cells, wherein the oral exfoliated cells are selected from the general population, and one side of the oral cavity is swabbed 20 times with a cotton swab;
[0043] DNA extraction: Use the Tiangen Biochemical DNA Extraction Kit to extract the genomic DNA of the cells, and strictly follow the instructions of the kit to extract the ...
specific Embodiment 2
[0074] It is illustrated by detecting the SNP site of the male AMELY gene homologous to the female AMELX gene. There are two genotypes at this site: male genotype CT and female genotype CC.
[0075] The selected samples come from: oral exfoliated cells, wherein the oral exfoliated cells are selected from company employees, and one side of the oral cavity is swabbed 20 times with a cotton swab;
[0076] DNA extraction: Use the Tiangen Biochemical DNA Extraction Kit to extract the genomic DNA of the cells, and strictly follow the instructions of the kit to extract the genomic DNA;
[0077] The position and flanking sequence of the SNP site to be detected in the reference genome (the reference genome version is hμManGRCh38 / hg38, see Table 6 below:
[0078] Table 6
[0079]
[0080] The corresponding primer probe sequences are shown in Table 7:
[0081] Table 7:
[0082] Numbering base sequence Remark SEQ 5 GGATGGCTGCACCACCAAATC XY Chromosome Common Pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com