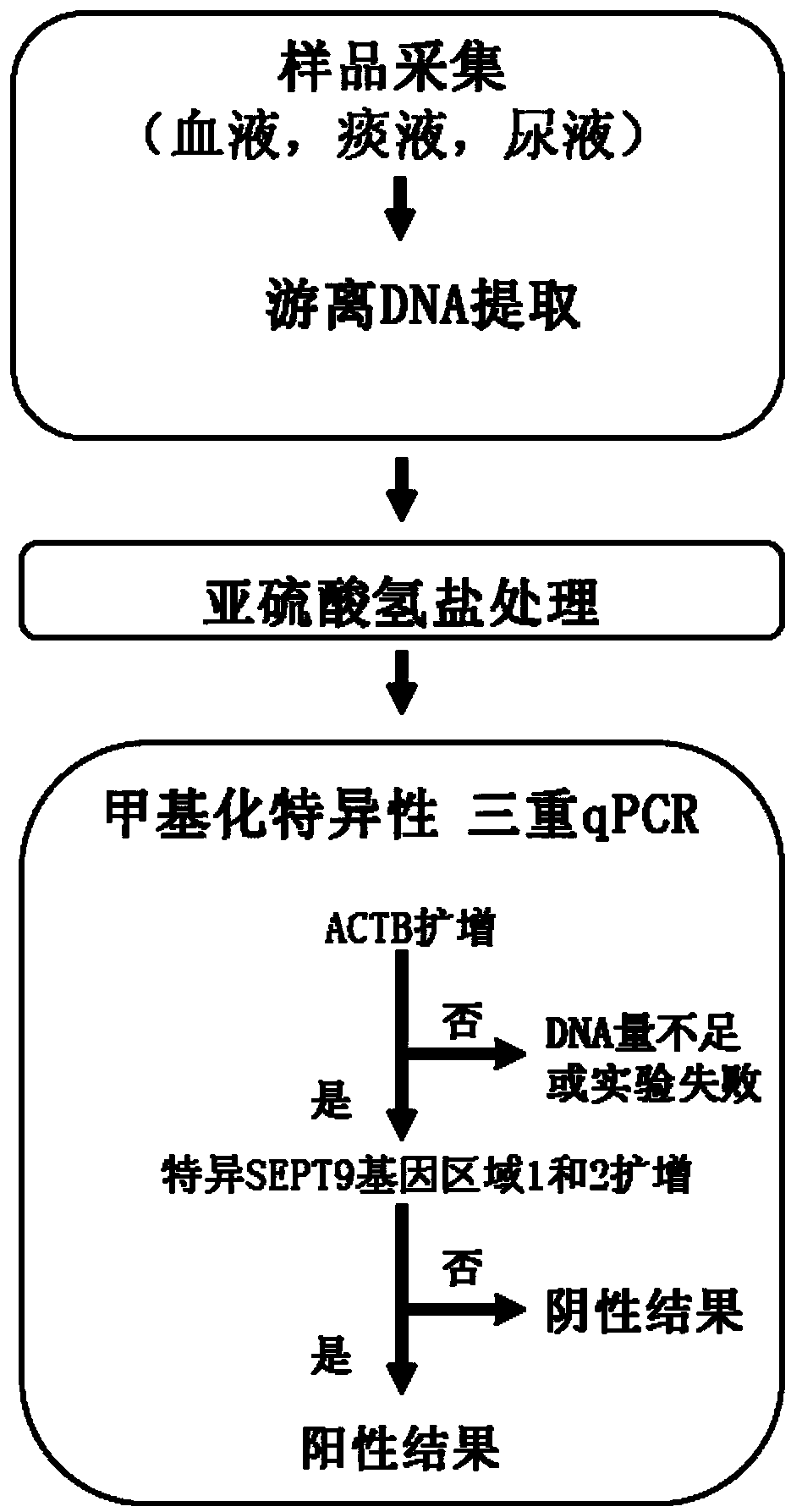
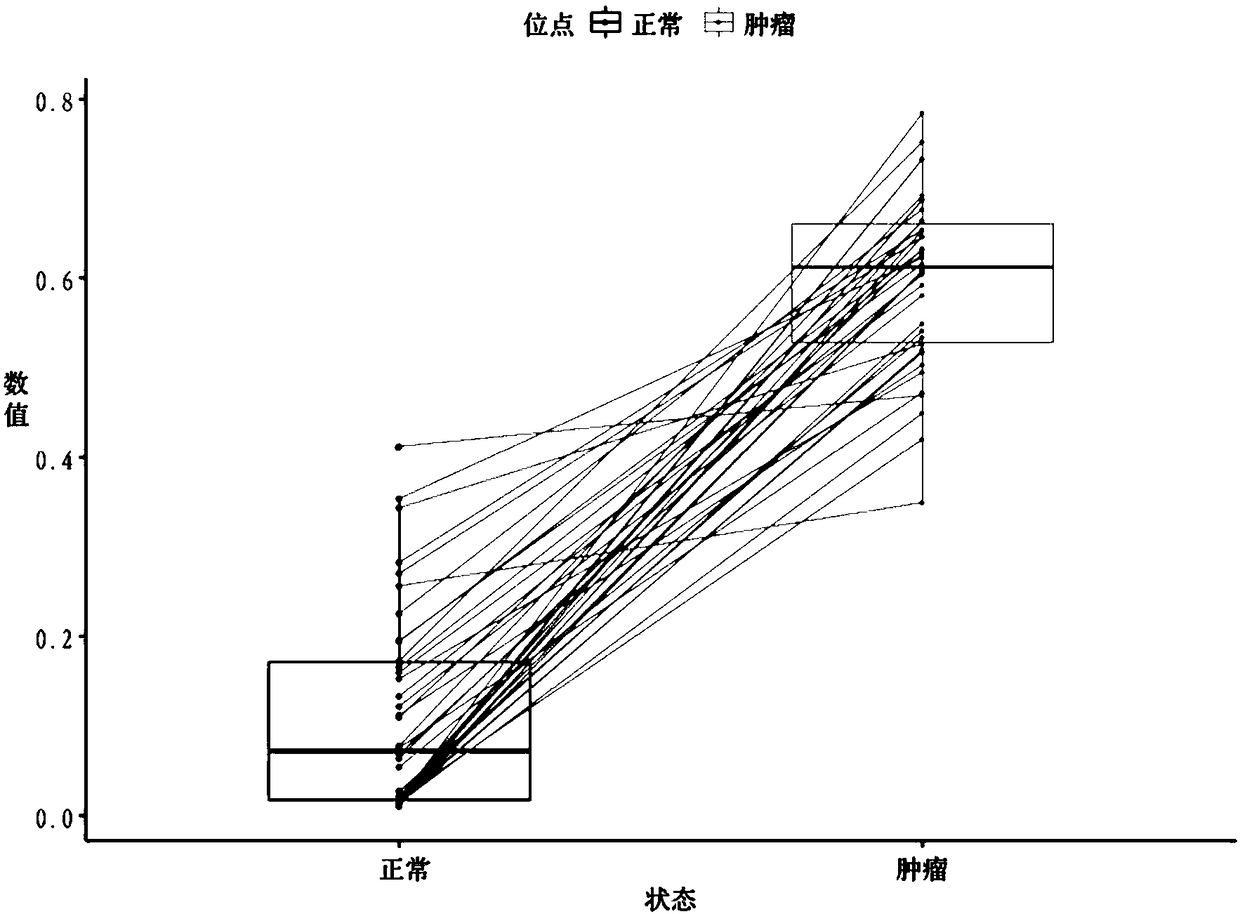
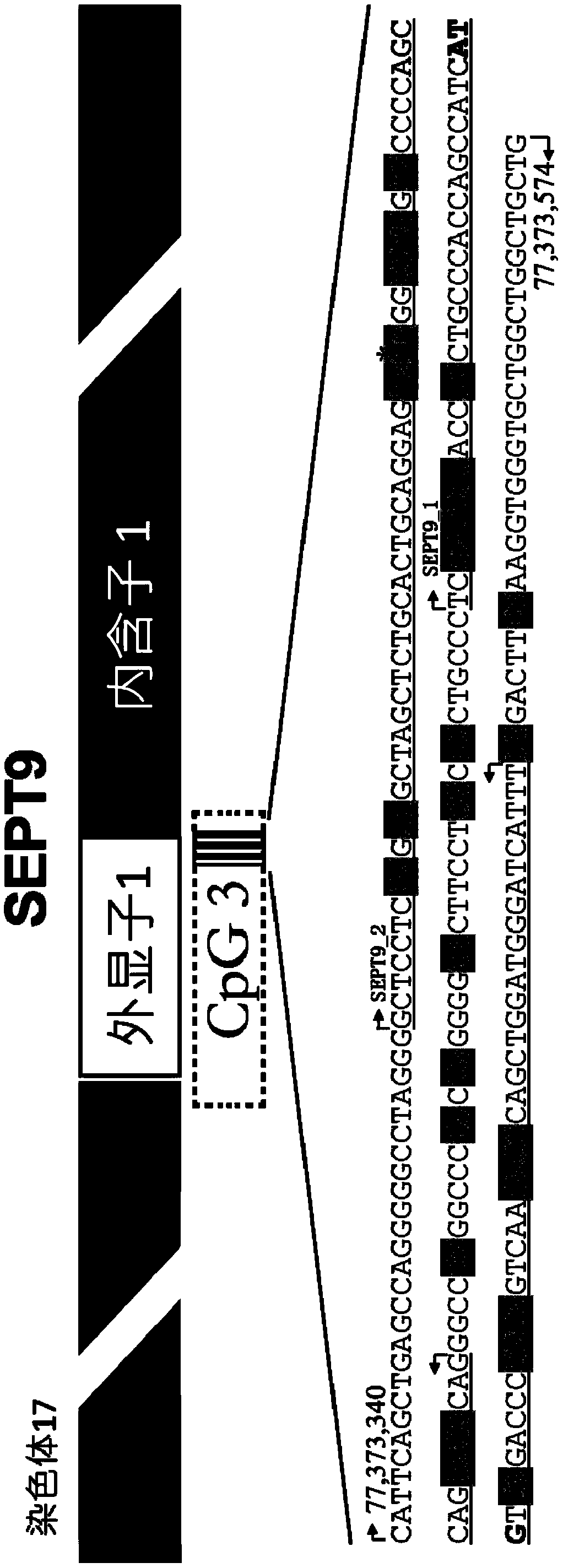
Kit and method for detecting DNA methylation by triplex qPCR assay
A kit and methylation technology, applied in the field of medicine and biology, can solve problems such as misdiagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1 SEPT9 gene methylation triple detection method sensitivity test
[0100] Experimental Materials
[0101] 1. Methylated human genomic DNA (purchased from NEB Company, item number N4007)
[0102] 2. EZ DNA Methylation-Lightning Kit (purchased from Zymo Research, Cat. No. D5031)
[0103] 3. Primers and probes corresponding to SEPT9 amplicon 1 (SEPT9_1) and amplicon 2 (SEPT9_2) (see the table below)
[0104] 4. ACTB amplification primers and probes (see the table below)
[0105] 5. AmpliTaq Gold TM DNA polymerase and buffer reagent (purchased from Thermo Fisher, Cat. No. 4311806)
[0106]
[0107] Wherein, the two nucleotides at the 3' end of each of the above primers are modified by thionucleotide; the DNA probe contains a TaqMan fluorophore at the 5' end, and a fluorescent quencher is attached to the 3' end.
[0108] experimental method
[0109] 1. DNA fragmentation treatment
[0110] In order to simulate the length of free DNA fragments, this experime...
Embodiment 2
[0123] Example 2 Comparison of SEPT9 gene methylation triple detection method with epiProcolon v2.0 high-sensitivity PCR kit
[0124] Experimental Materials
[0125] 1. Unmethylated human genomic DNA (purchased from Promega, Cat. No. G3041)
[0126] 2. Methylated human genomic DNA (purchased from NEB Company, item number N4007)
[0127] 3. EZ DNA Methylation-Lightning kit (purchased from Zymo Research, Cat. No. D5031)
[0128] 4. SEPT9 amplicon 1 (SEPT9_1) and amplicon 2 (SEPT9_2) corresponding primers and probes (same as Example 1)
[0129] 5.ACTB amplification primer and probe (with embodiment 1)
[0130] 6. AmpliTaq Gold TM DNA polymerase and buffer reagent (purchased from Thermo Fisher, Cat. No. 4311806)
[0131] 7. epiProcolon v2.0 High Sensitive PCR Kit (purchased from Epigenomics, Cat. No. M5-02-002)
[0132] experimental method
[0133] 1. DNA fragmentation treatment
[0134] The non-methylated human genomic DNA and the methylated human genomic DNA are ultraso...
Embodiment 3
[0151] Example 3 Detection of normal control samples using SEPT9 methylation triple qPCR detection and epiProcolon v2.0 high-sensitivity PCR kit
[0152] Experimental Materials
[0153] 1. Healthy human plasma (purchased from Bloodworks NW company)
[0154] 2. QIAamp Circulatory System Nucleic Acid Extraction Kit (purchased from Qiagen, Cat. No. 55114)
[0155] 3. EZ DNA Methylation-Lightning kit (purchased from Zymo Research, Cat. No. D5031)
[0156] 4. SEPT9 amplicon 1 (SEPT9_1) and amplicon 2 (SEPT9_2) corresponding primers and probes (same as Example 1)
[0157] 5.ACTB amplification primer and probe (with embodiment 1)
[0158] 6. AmpliTaq Gold TM DNA polymerase and buffer reagent (purchased from Thermo Fisher, Cat. No. 4311806)
[0159] 7. epiProcolon v2.0 High Sensitive PCR Kit (purchased from Epigenomics, Cat. No. M5-02-002)
[0160] experimental method
[0161] 1. DNA extraction from plasma
[0162] Cell-free DNA was extracted from 10 plasma samples using the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com