Preparation method of D-amino acid
An amino acid and acetyl amino acid technology, applied in biochemical equipment and methods, enzymes, hydrolase and other directions, can solve the problems of low product concentration and high production cost, achieve high yield, good product quality, and reduce the cost of enzymes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1. Construction of recombinant bacteria
[0032] clone derived from Aspergillus oryzae The N-acetyl-D-aminoacylase gene was connected into pET41a vector (Novagen) or pKK223-3 vector, transformed into BL21(DE3) host bacteria, and D-acylase recombinant bacteria (NLase) were constructed.
[0033] clone derived from Sebekia Benihana The N-acetyl-amino acid racemase gene was connected to the pET41a vector (Novagen Company), and transformed into BL21(DE3) host bacteria to construct N-acetyl amino acid racemase recombinant bacteria (NAAR).
[0034] 2. Fermentation of recombinant bacteria to prepare enzymes
[0035] The recombinant bacteria NLase and NAAR were fermented with TB.
[0036] 1) culture medium
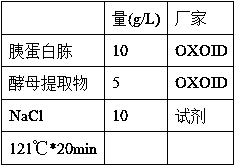
[0037] (1) LB:
[0038]
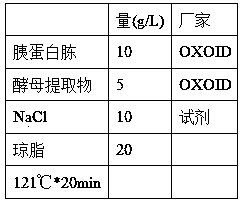
[0039] (2) Agar LB-Kan plate (g / L):
[0040]
[0041] Note: The culture medium is sterilized and cooled to 50-60°C, add 100μl Kan solution (100mg / ml) to 100ml LB, mix well, and pour it onto a plate (25-30ml / 6cm plate).
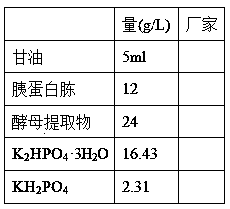
[0042] (3) TB...
Embodiment 2
[0060] 1. Construction of recombinant bacteria
[0061] N-acetyl-D-aminoacylase derived from Alcaligenes denitrificans was cloned, and the gene was connected to pKK223-3 vector (Novagen Company) or transformed into BL21(DE3) host bacteria to construct D-acylase recombinant bacteria (NLase).
[0062] The N-acetyl-amino acid racemase derived from Amycolatopsis azurea was cloned, the gene was connected into pET28a vector (Novagen), and transformed into BL21(DE3) host bacteria to construct N-acetyl amino acid racemase recombinant bacteria (NAAR).
[0063] 2. Fermentation of recombinant bacteria to prepare enzymes
[0064] The recombinant bacteria NLase and NAAR were fermented with TB.
[0065] 1) culture medium
[0066] With embodiment 1.
[0067] 2) Fermentation process
[0068] (1) Strain activation: inoculate the recombinant bacteria NLase and NAAR on Kan LB agar plates respectively, and culture them at 37°C for 12-14 hours.
[0069] (2) First-class seeds: Pick a single ba...
Embodiment 3
[0083] 1. Construction of recombinant bacteria
[0084] The N-acetyl-D-aminoacylase derived from Alcaligenes denitrificans was cloned, the gene was connected into pET28a vector (Novagen), and transformed into BL21(DE3) host bacteria to construct D-acylase recombinant bacteria (NLase).
[0085] The N-acetyl-amino acid racemase derived from Deinococcus radiodurans was cloned, the gene was connected into pET28a vector (Novagen), and transformed into BL21(DE3) host bacteria to construct N-acetyl amino acid racemase recombinant bacteria (NAAR).
[0086] 2. Fermentation of recombinant bacteria to prepare enzymes
[0087] The recombinant bacteria NLase and NAAR were fermented with TB.
[0088] 1) culture medium
[0089] With embodiment 1.
[0090] 2) Fermentation process
[0091](1) Strain activation: Inoculate the recombinant bacteria NLase and NAAR on Kan LB agar plates respectively, and culture them at 37°C for 14 hours.
[0092] (2) First-class seeds: Pick a single bacterium...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com