Nucleic acid aptamer and its application in the detection of neuronecrosis virus derived from pompano ovata
A nucleic acid aptamer and nucleotide sequence technology, which can be applied to measurement devices, instruments, biochemical equipment and methods, etc., can solve the problems of inability to detect and diagnose the oval pomfret-derived nerve necrosis virus, and achieve convenient in vitro chemical Synthetic, easy to label, small molecular weight effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 The preparation method of SSDNA nucleic acid adapter is as follows:
[0038] Step 1: Single-stranded DNA libraries and primers shown in the following sequences:
[0039] Random library library50:
[0040] 5'-gtctgaagtagacgcaggag (50n) agtcacctgagtaagcgt
[0041] 5 'primers: 5'-fam-gtctgaagtagacgcaggag-3';
[0042] 3 'primers: 5'-biotin-acgcttactcaggtgtgact-3';
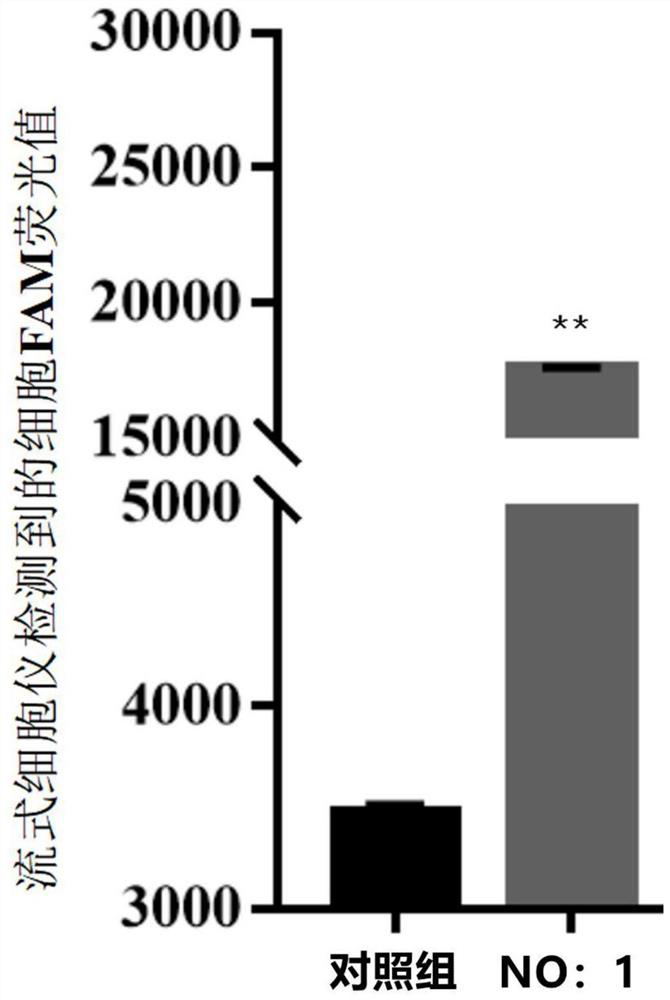
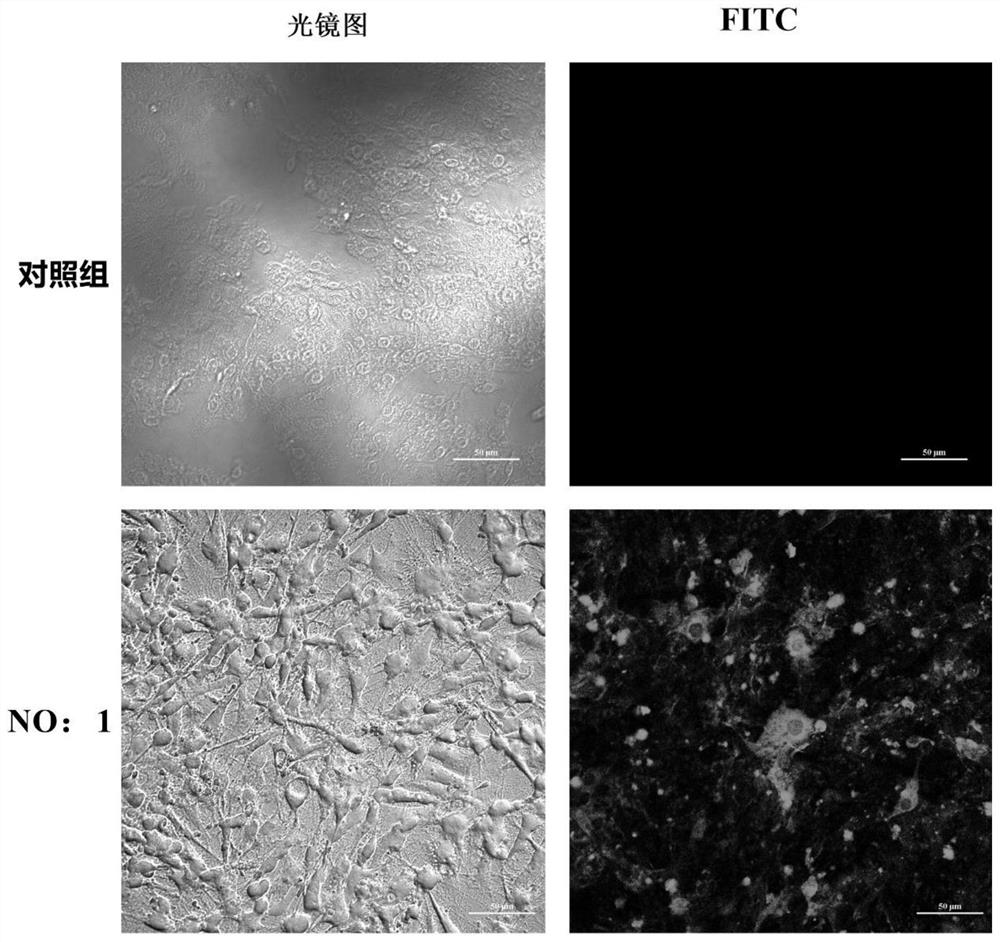
[0043] Step 2: Take 10 nmol The random library is dissolved in 500 μl of PBS, 5 min at 92 ° C, then quickly ice bath, ice bath 10min, and take the treated random library and GTONNV infected cells to incubate on ice 1h; After the completion of the combination, the supernatant was centrifuged, and 10 ml of PBS was washed with GTONNV infected cells, and 10 min at a constant temperature water bath at 92 ° C and centrifuged at 12000 g of 1-20 min, and the supernatant was collected, the supernatant SSDNA nucleic acid adapter library for specific recognition of GTONNV infected cells;
[0044] Step 3: Take 100 ul ...
Embodiment 2
[0055] Example 2 The preparation method of SSDNA nucleic acid adapter is as follows:
[0056] Step 1: Single-stranded DNA libraries and primers shown in the following sequences:
[0057] Random library library50:
[0058] 5'-gtctgaagtagacgcaggag (50n) agtcacctgagtaagcgt
[0059] 5 'primers: 5'-fam-gtctgaagtagacgcaggag-3';
[0060] 3 'primers: 5'-biotin-acgcttactcaggtgtgact-3';
[0061] Step 2: Take 10 nmol The random library is dissolved in 500 μl of PBS, 5 min at 92 ° C, then quickly ice bath, ice bath 10min, and take the treated random library and GTONNV infected cells to incubate on ice 1h; After the completion of the combination, the supernatant was centrifuged, and 10 ml of PBS was washed with GTONNV infected cells, and 10 min at a constant temperature water bath at 92 ° C and centrifuged at 12000 g of 1-20 min, and the supernatant was collected, the supernatant SSDNA nucleic acid adapter library for specific recognition of GTONNV infected cells;
[0062] Step 3: Take 100 ul ...
Embodiment 3
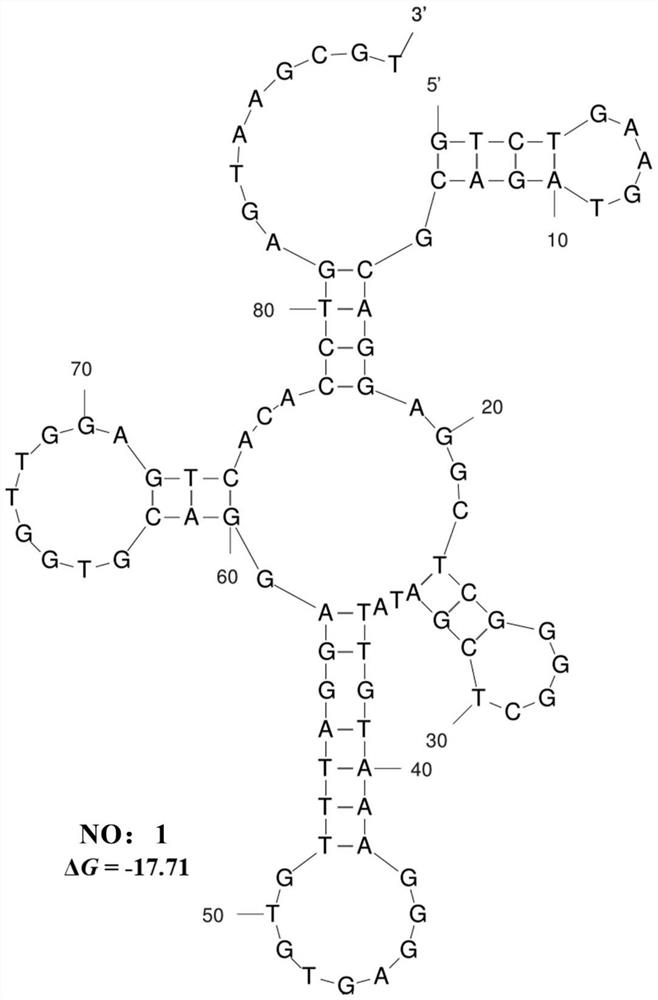
[0085] The secondary structure of the nucleic acid fitting is predicted online with Mfold software online.
[0086] SEQ ID NO: 1 secondary structural prediction result of nucleic acid body fit image 3 As shown, the nucleic acid adapter forms a special stem ring structure and a cardboard structure.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com