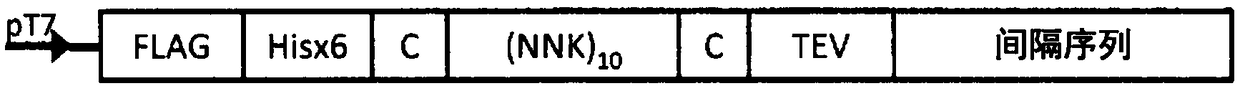Ribosome display complex and production method therefor
A technology of ribosome display and manufacturing method, which is applied in the field of ribosome display complex and its manufacturing, and can solve the problems of difficult technology and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0104] (1) Preparation process of ribosome display complex
[0105] In this step, mRNA molecules are translated using a cell-free peptide synthesis system using ribosomes to obtain ribosome complexes including unmodified polypeptide chains, mRNA molecules, and ribosomes.
[0106] The cell-free peptide synthesis system using ribosomes is a system for synthesizing polypeptides from mRNA in vitro using compounds required for polypeptide synthesis based on RNA information carried out in cells. Specifically, mRNA molecules are added to a reaction system containing proteins, ribosomes, tRNA, amino acids, NTP, buffers, etc. required for mRNA translation, energy regeneration, etc. , to synthesize a polypeptide corresponding to the added mRNA. Since a kit for synthesizing a cell-free peptide is commercially available, it is sufficient to use a commercially available kit other than the mRNA molecule.
[0107] The ribosome display complex prepared in this step includes mRNA, a polypept...
Embodiment 1
[0165] (1) Preparation of RNA library
[0166] In this (1) project, for the use of NNK method to make 10 12 The method of the RNA library of the above RNA is described, the RNA has a (NNK) comprising (NNK) 10 [wherein, N represents A, U, G or C, K represents G or U, and NNK corresponds to all codons] the sequence of the sequence.
[0167] To make this RNA library, use a figure 1 The structure of the template DNA (base sequence: SEQ ID NO: 1, amino acid sequence: SEQ ID NO: 2). Specifically, using the reaction solution having the composition shown in Table 1, a 5' fragment was prepared using the plasmid as a template DNA in the PCR cycle in Table 2. In Table 1, 5FFnew_130816 is the forward primer, and Ma3frag_R0502 is the reverse primer.
[0168] [Table 1]
[0169]
[0170] [Table 2]
[0171]
[0172] Next, using the reaction solution having the composition shown in Table 3, the 3' fragment of the template DNA was prepared according to the PCR cycle in Table 4. In...
Embodiment 2
[0211] Embodiment 2: the cyclization reaction of peptide
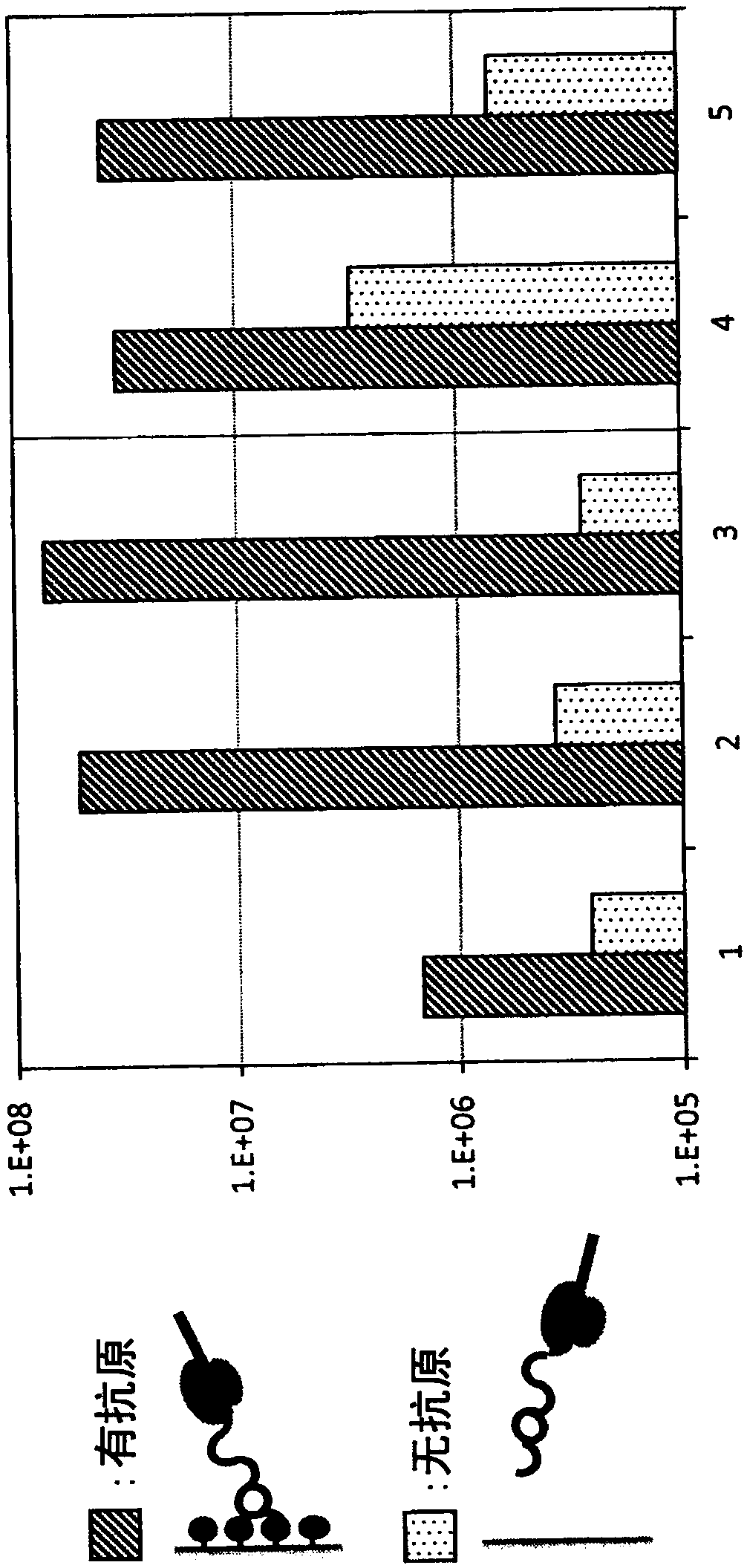
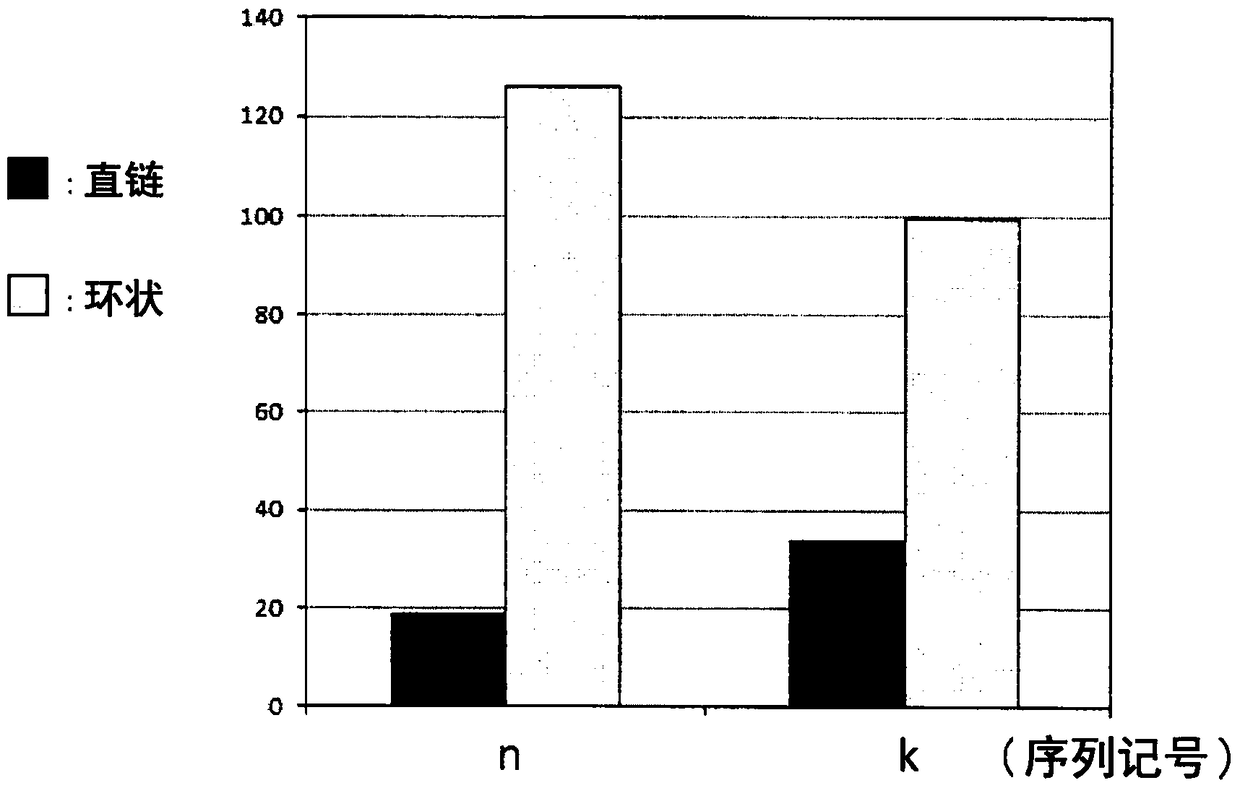
[0212] Using a recombinant cell-free protein synthesis kit ("PURE frex (registered trademark)" manufactured by GeneFrontier Corporation), RNA having a FLAG sequence, a His6 sequence, and a nucleotide sequence encoding a TEV protease site (SEQ ID NO. 8) 2.5×10 13 The molecules were reacted at 37°C for 35 minutes to prepare an RD complex, and anti-FLAG (registered trademark) M2 antibody-bound agarose beads (manufactured by Sigma-Aldrich, 2 μL) were added to the reaction solution to bind to the RD complex. Further, tris(2-carboxyethyl) sodium salt (pH 7, final concentration 0.5 mM) was added as a reducing agent and Figure 4 Each modification reagent at the indicated final concentration of 2 mM was stirred at 4° C. for 3 hours, thereby cyclizing the peptide in the RD complex on the beads. After the reaction, the RD complex was released from the beads by adding FLAG peptide (sequence: DYKDDDDK, 5 mg). Separate and remov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com