Sucrose hydrolase mutant as well as preparation method and application of sucrose hydrolase mutant
A technology of sucrose hydrolase and mutants, applied in the field of genetic engineering and enzyme engineering, to achieve the effect of improving the activity of transglycosides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1: Recombinant bacteria construction
[0034] According to the sucrose hydrolase gene sequence whose accession number is YP_002516566.2 on NCBI, the CcSH gene containing sucrose hydrolase was synthesized by chemical synthesis, and the sucrose hydrolase gene sequence whose accession number was CDG80999.1 was synthesized by chemical synthesis. JaSH gene, accession number is the sucrose hydrolase gene sequence of AAQ93678.1 The XaSH gene containing sucrose hydrolase is synthesized by chemical synthesis. CcSH gene, JaSH gene and XaSH gene were digested with pET-24a(+) plasmid with NdeI and HindⅢ, respectively, and the digested products were recovered by cutting gel, then ligated with T4 ligase, and the ligated products were transformed into E.coli JM109 competent cells , to obtain recombinant cells. The recombinant cells were cultured at 37°C for 8 hours, and the transformants were picked and cultured with shaking in LB liquid medium (containing 30mg / L kanamycin...
Embodiment 2
[0037] Embodiment 2: the preparation of sucrose hydrolase mutant
[0038] (1) Preparation of single mutation of sucrose hydrolase
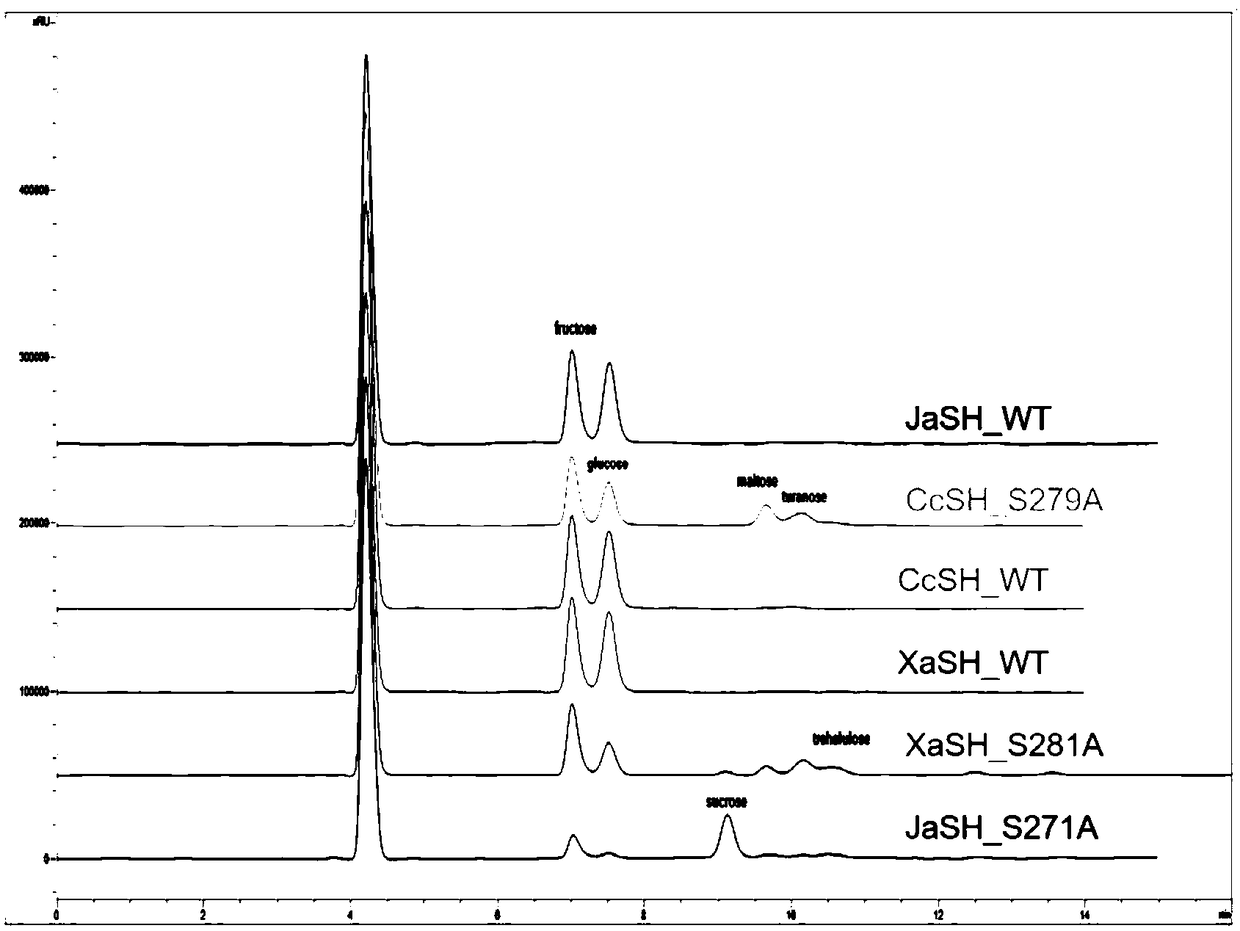
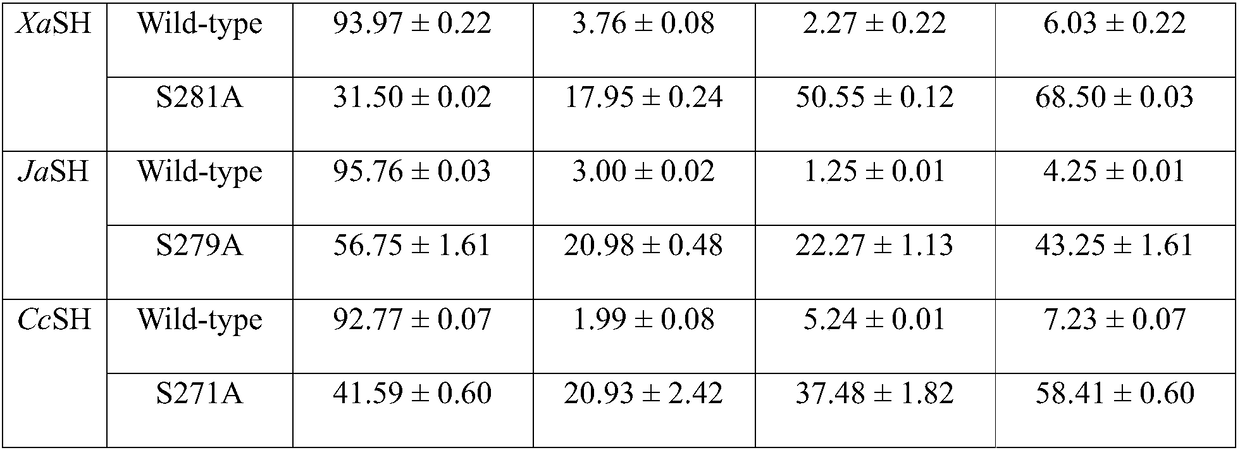
[0039] According to the CcSH gene sequence of sucrose hydrolase, primers for introducing the S271A mutation were designed and synthesized, and the plasmid CcSH / pET-24a(+) carrying the gene encoding wild-type sucrose hydrolase was used as a template by rapid PCR technology to detect sucrose hydrolase Site-directed mutation was performed on the CcSH gene sequence, and the DNA coding sequence was determined to identify the gene in which the 271st Ser codon was changed to Ala codon, and the sucrose hydrolase single mutation S271A was obtained.
[0040] According to the JaSH gene sequence of sucrose hydrolase, primers for introducing the S279A mutation were designed and synthesized. Using rapid PCR technology, the plasmid JaSH / pET-24a(+) carrying the gene encoding wild-type sucrose hydrolase was used as a template to target sucrose hydrolase. The JaSH...
Embodiment 3
[0063] Embodiment 3: the concentration of crude enzyme liquid
[0064] Slowly add ammonium sulfate with a concentration of 20% relative to the mass fraction of the enzyme liquid to the enzyme liquid obtained in Examples 1 and 2 while stirring, stir until the ammonium sulfate is dissolved, and stand at 4°C for 8-10 hours to precipitate protein. The mixture was centrifuged (8000rpm, 10min) to collect the precipitate, and the minimum volume of 50mM KH 2 PO 4 -Na 2 HPO 4 The buffer (pH 7.0) was redissolved, and after reconstitution, the solid matter was removed by centrifugation again, and the supernatant was collected and dialyzed to obtain a concentrated enzyme solution.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com