Molecular marker primer for controlling QTL locus of corn ear length and application
A technology of molecular markers and ears of corn, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of slow, less reported progress, rare reports of ear length sites, etc., and achieve good results Breeding value, effect of increasing ear length
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Development of Molecular Markers Tightly Linked to Major QTL for Ear Length in Maize:
[0044] 1 Materials and methods
[0045] 1.1 Experimental materials
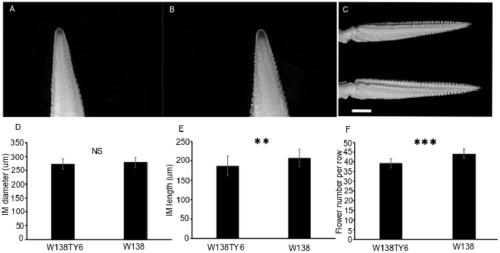
[0046] Maize Inbred Line W138, ZmEL-1 Near Isogenic Line W138 TY6 and the progeny mapping population constructed by the near isogenic line.
[0047] 1.2 Test method
[0048] 1.2.1 Target segment molecular marker development
[0049] According to the B73 genome, download the ZmEL-1 interval sequence, use the SSRSCAN tool of the REDB website to search for the SSR sequence, and then design specific primers based on the SSR flanking sequence, and then perform amplification based on the parent material W138 and TY6, and perform capillary electrophoresis Perform polymorphism analysis, and select markers with polymorphism and clear bands for population genotype analysis.
[0050] 1.2.2 Field experiments and identification of phenotypes
[0051] For the heterozygous recombinant single plant used for the offspring test...
Embodiment 2
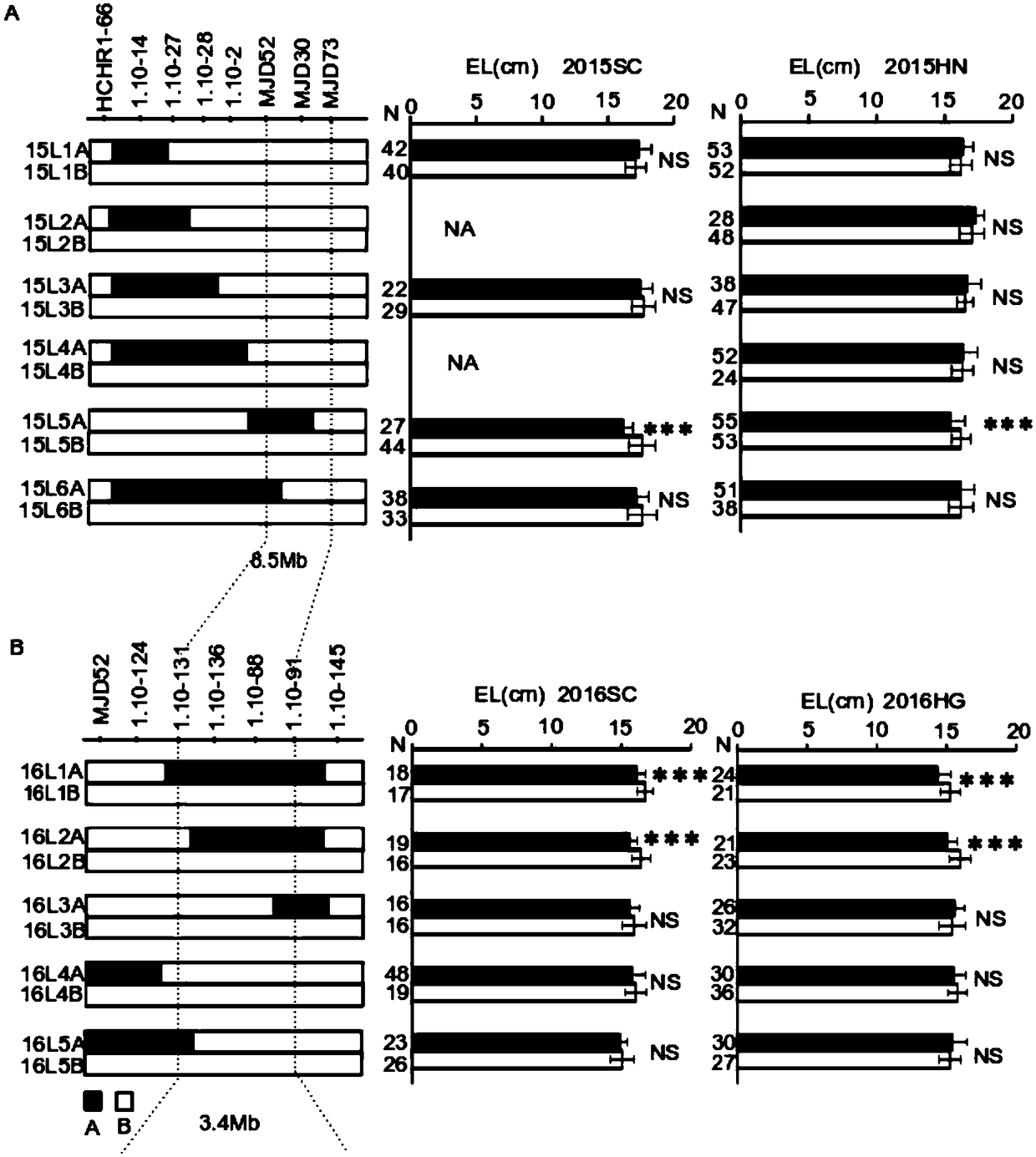
[0088] The application of molecular marker primers closely linked to the major QTL ZmEL-1 of maize ear length in maize breeding:
[0089] 1. Materials and methods
[0090] 1.1 Experimental materials
[0091] Maize inbred line W138 and its near isogenic line W138 TY6 , 9 excellent corn inbred lines: HL78F (the male inbred line of Hualiang 78 cultivated by Hualiang Seed Industry), 501, HZ4, SHENG62 (the male inbred line of Shengrui 999 cultivated for Shengrui Company) , CHANG7-2, YE478, ZHENG58, 4CV, 6WC.
[0092] 1.2 Test method
[0093] 1.2.1 ZmEL-1 near-isogenic line W138 and its near-isogenic line W138 were selected based on molecular markers TY6 Hybrid combination with excellent inbred lines
[0094] Nine elite maize inbred lines (HL78F, 501, HZ4, SHENG62, CHANG7-2, YE478, ZHENG58, 4CV, 6WC) derived from different heterosis groups of maize were selected and selfed with the superior ZmEL-1 genotypes respectively. Line W138 (using molecular markers 1.10-131, 1.10-91 for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com