Amplification primers, screening methods and identification methods for identification of olive varieties based on SNP sites
A technology for amplification primers and species identification, which is applied in biochemical equipment and methods, microbiological determination/inspection, recombinant DNA technology, etc., can solve the problems of various products, chaotic peak shapes, and expensive SSR detection, etc., to achieve expansion High efficiency, clear peak shape, low cost detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] In 2016, the Forestry Research Institute of the Chinese Academy of Forestry collected an olive variety resource A from the Wudu olive planting base in Gansu. There is still a difference. In order to identify whether the variety resource A is 'Dou Guo', the following processing analysis was carried out:
[0048] 1. Collect the leaf materials of olive variety A and the known variety 'Douguo' respectively, and extract DNA;
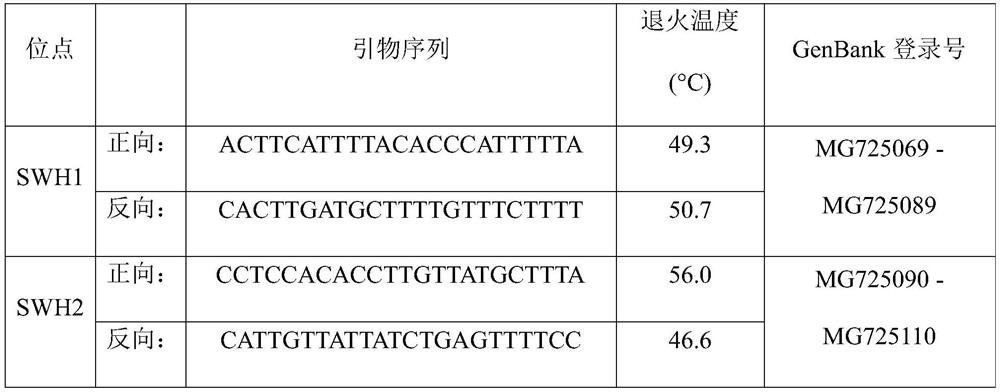
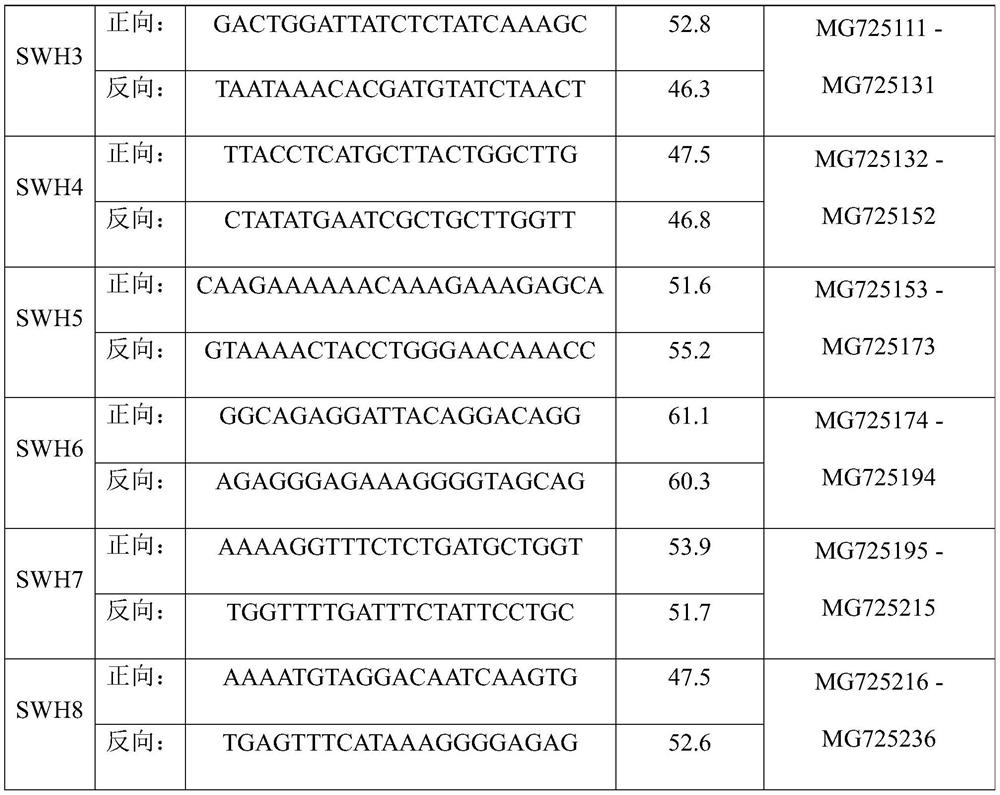
[0049] 2. Use 8 pairs of olive amplification primers in Table 1 of the present invention to amplify and sequence the two DNAs respectively;
[0050] PCR amplification uses 30uL reaction system: 20ng DNA, 10×buffer (Promega, USA) buffer 3uL, 2.4mM dNTPs 2.4uL, 10uM primer 2.4uL, forward primer and reverse primer 1.2uL each, TaqDNA polymerase (5U / uL, Takara, Shiga, Japan) 0.15uL. ABI 96U Thermo cycler PCR instrument was used for PCR amplification.
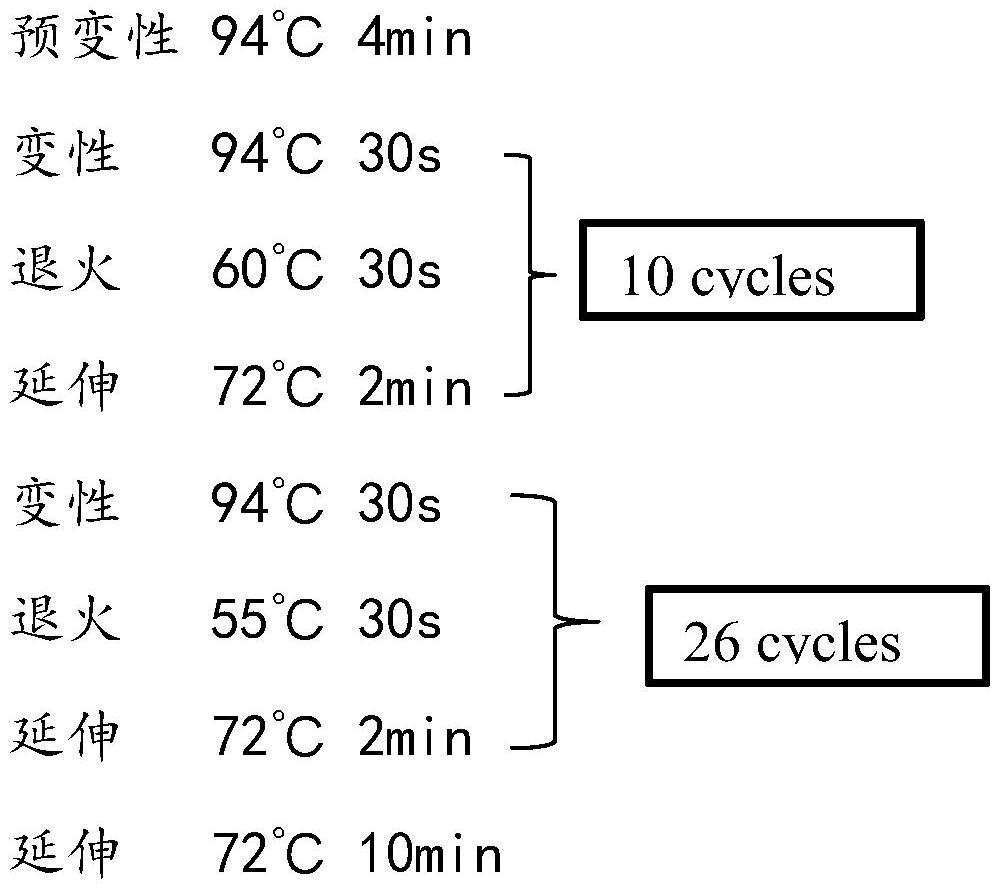
[0051] The reaction procedure of PCR amplification is:
[0052]
[0053] 3. Use ContigExpress soft...
Embodiment 2
[0057] In 2016, the Forestry Research Institute of the Chinese Academy of Forestry collected an olive variety resource B from the olive planting base in Mianyang, Sichuan. Since there are no important varieties such as fruits, it is not clear what variety it is. To identify this variety resource B, the following treatment analyzes were performed:
[0058] 1. Collect the leaf material of olive variety B and extract DNA;
[0059] 2. Use 8 pairs of olive amplification primers in Table 1 of the present invention to amplify and sequence the leaf DNA of this variety resource respectively;
[0060] PCR amplification uses 30uL reaction system: 20ng DNA, 10×buffer (Promega, USA) buffer 3uL, 2.4mM dNTPs 2.4uL, 10uM primer 2.4uL, forward primer and reverse primer 1.2uL each, TaqDNA polymerase (5U / uL, Takara, Shiga, Japan) 0.15uL. ABI 96U Thermo cycler PCR instrument was used for PCR amplification.
[0061] The reaction procedure of PCR amplification is:
[0062]
[0063] 3. Use Co...
Embodiment 3
[0068] In 2017, the Subtropical Forestry Research Institute of the Chinese Academy of Forestry collected an olive variety resource C from the Lishui olive planting base in Zhejiang. The resource of this variety was introduced from Sichuan, and its phenotypic traits are similar to the existing olive variety 'Leixing', but there are still differences in traits such as leaf texture. In order to identify whether the variety resource C is 'Leixing', the following processing analysis was carried out:
[0069] 1. Collect the leaf material of olive variety C and extract DNA;
[0070] 2. Use 8 pairs of olive amplification primers in Table 1 of the present invention to amplify and sequence the leaf DNA of this variety resource respectively;
[0071] PCR amplification uses 30uL reaction system: 20ng DNA, 10×buffer (Promega, USA) buffer 3uL, 2.4mM dNTPs 2.4uL, 10uM primer 2.4uL, forward primer and reverse primer 1.2uL each, TaqDNA polymerase (5U / uL, Takara, Shiga, Japan) 0.15uL. ABI 96...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com