Kit for non-invasive prenatal screening for trisomy syndrome and application thereof
A technique for trisomy syndrome and trisomy 21, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve the problems of low detection sensitivity, high cost of next-generation sequencing technology, detection Long process and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1. Screening of trisomy-specific repeat sequences and detection primers and probes thereof
[0082] 1. Screening of trisomy-specific repeat sequences
[0083] 1. Database normalization
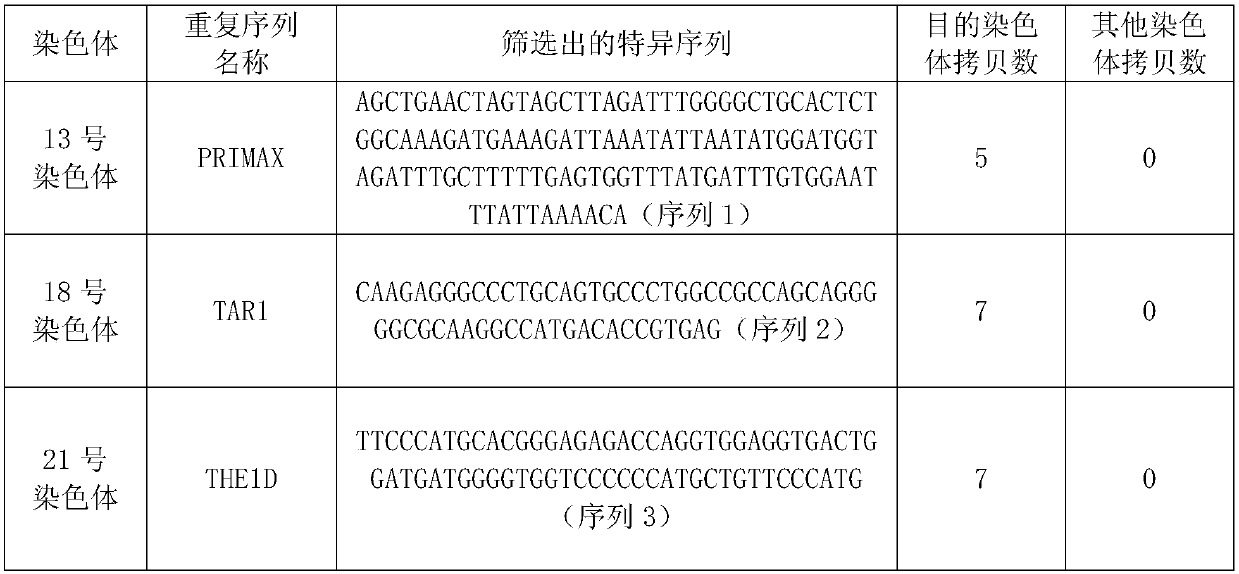
[0084] Download all the repetitive sequences in the human genome sequence hg38 from the repeatmaster software, and extract the chromosomes, chromosome start and end positions, repeat sequence names, repeat sequence start and end positions, and repeat sequence types according to its unique format As well as the base sequence of the repetitive sequence, it is integrated into the fasta format, and the standardized database is obtained and saved.
[0085] 2. Screening of feature sequences
[0086] In the normalized database, search for specific repetitive sequences of more than 50 bp in the two regions, which need to have a copy number > 5, and there is no variation or polymorphism. These repetitive sequences are searched outside the region in turn, and if there is the same sequen...
Embodiment 2、18 and 21 3
[0097] The detection method of embodiment 2, 18 and 21 trisomy
[0098] 1. Preparation of samples to be tested
[0099] 1. Experimental group samples
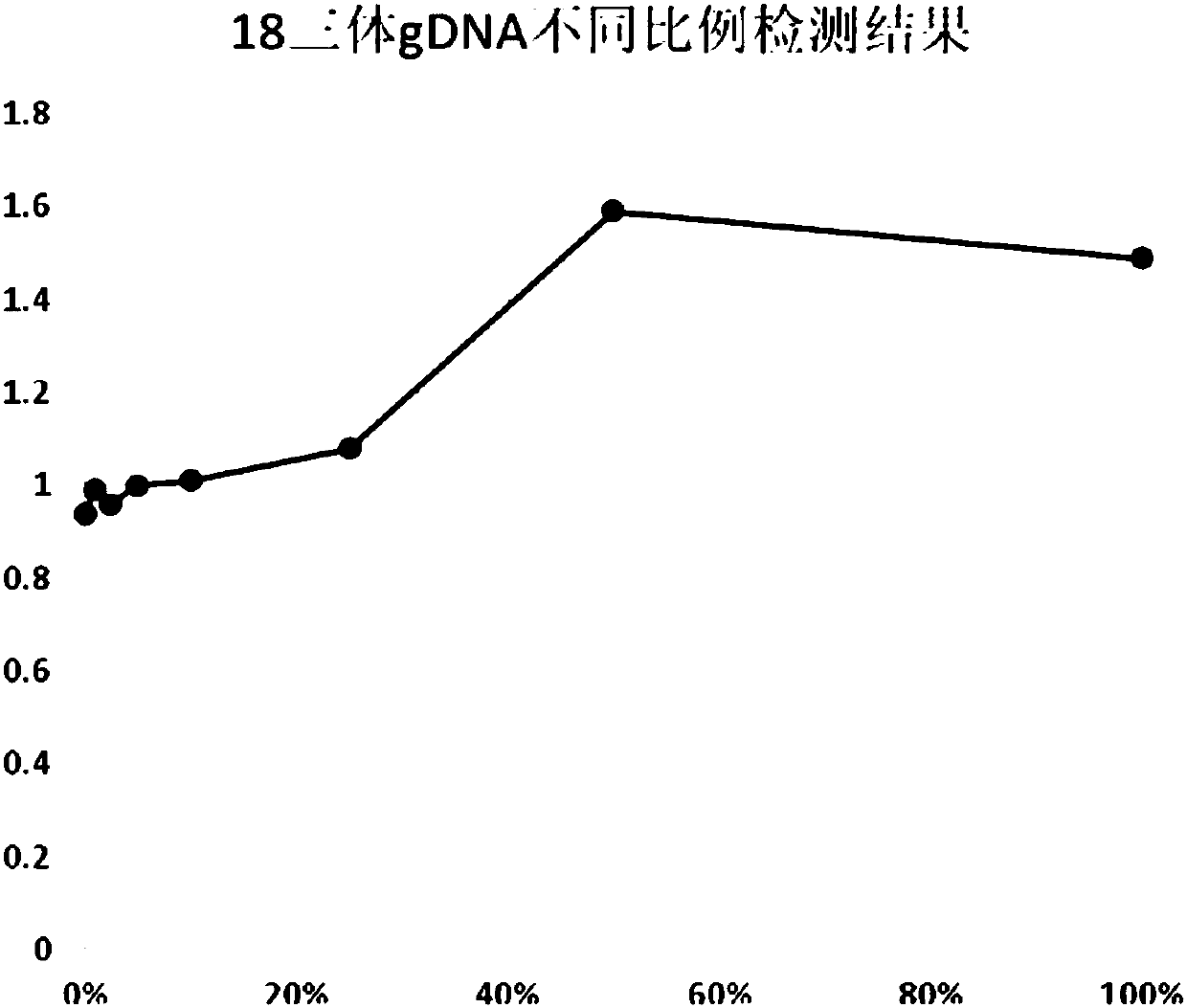
[0100] (1) Trisomy 18 DNA (DNA of patients with clinically diagnosed trisomy 18 syndrome), YH cell line DNA, and mixed samples of trisomy 18 DNA and YH cell line DNA were used as samples to be tested. The mixed samples of trisomy 18 DNA and YH cell line DNA are mixed according to the following proportions: 50%, 25%, 10%, 5%, 2.5%, 1% and 0.5% to obtain trisomy 18 with different mutation frequencies Mixed samples. The DNA concentration in each sample to be tested is 2ng / ul.
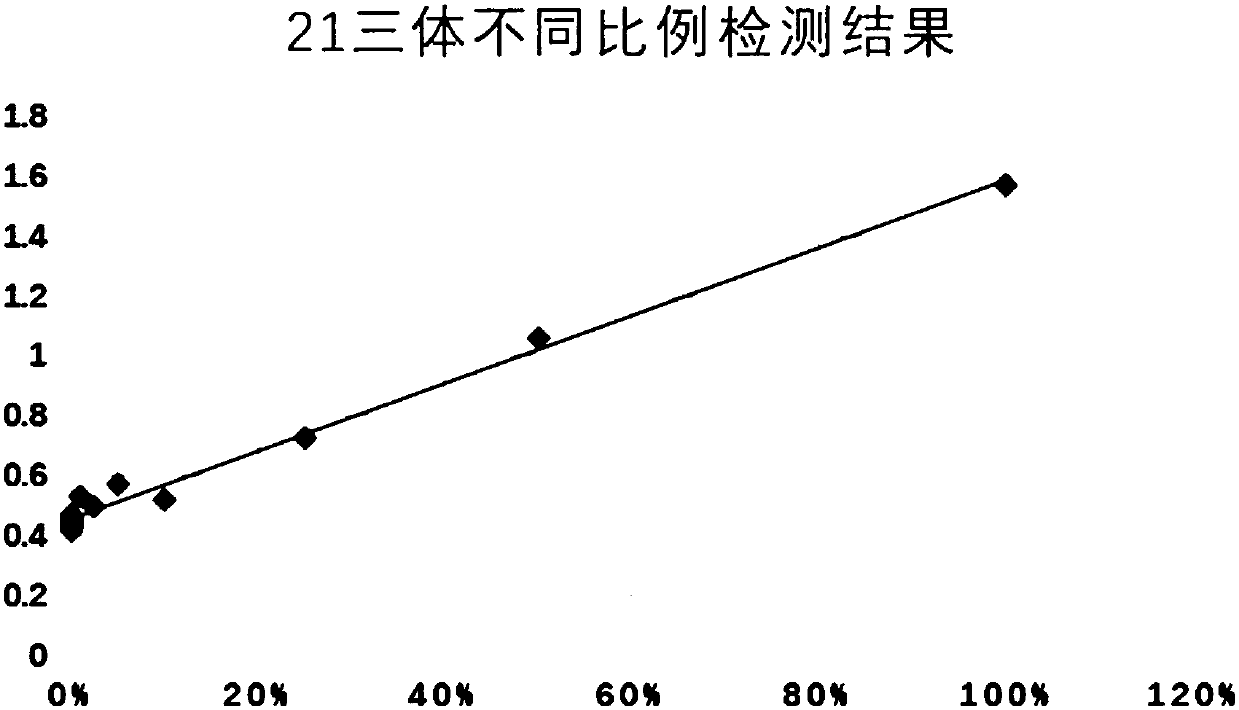
[0101] (2) Trisomy 21 DNA (DNA of a clinically diagnosed trisomy 21 patient), YH cell line DNA, and a mixed sample of trisomy 21 DNA and YH cell line DNA were used as samples to be tested. The mixed samples of trisomy 21 DNA and YH cell line DNA are mixed according to the following proportions: 50%, 25%, 10%, 5%, 2.5%, 1% and 0.5% to obtain trisomy 21 w...
Embodiment 3
[0141] The specific detection of embodiment 3, primer and probe
[0142] (1) Specific detection of primers
[0143] 1. Using normal human cell DNA as a template, PCR amplification was performed using the PRIMAX-F and PRIMAX-R primers designed in Example 1 to obtain a PCR amplification product.
[0144] The above PCR amplification products were detected by agarose gel electrophoresis. The results showed that a band with a size of about 60bp was obtained by PCR amplification, which was consistent with the expectation and the band was single, indicating that the primers of PRIMAX-F and PRIMAX-R had good specificity.
[0145] 2. Using normal human cell DNA as a template, PCR amplification was performed using the TAR-F and TAR-R primers designed in Example 1 to obtain a PCR amplification product.
[0146] The above PCR amplification products were detected by agarose gel electrophoresis. The results showed that a band of about 60 bp was obtained by PCR amplification, which was co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com