In vivo plasmid editing system based on CRISPR/Cas and lambda-Red recombination system, and application thereof
An editing system and plasmid technology, applied in microorganism-based methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve problems such as low efficiency and limitations of recombination, and achieve the effects of short time period, reduced cost, and expanded scope of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
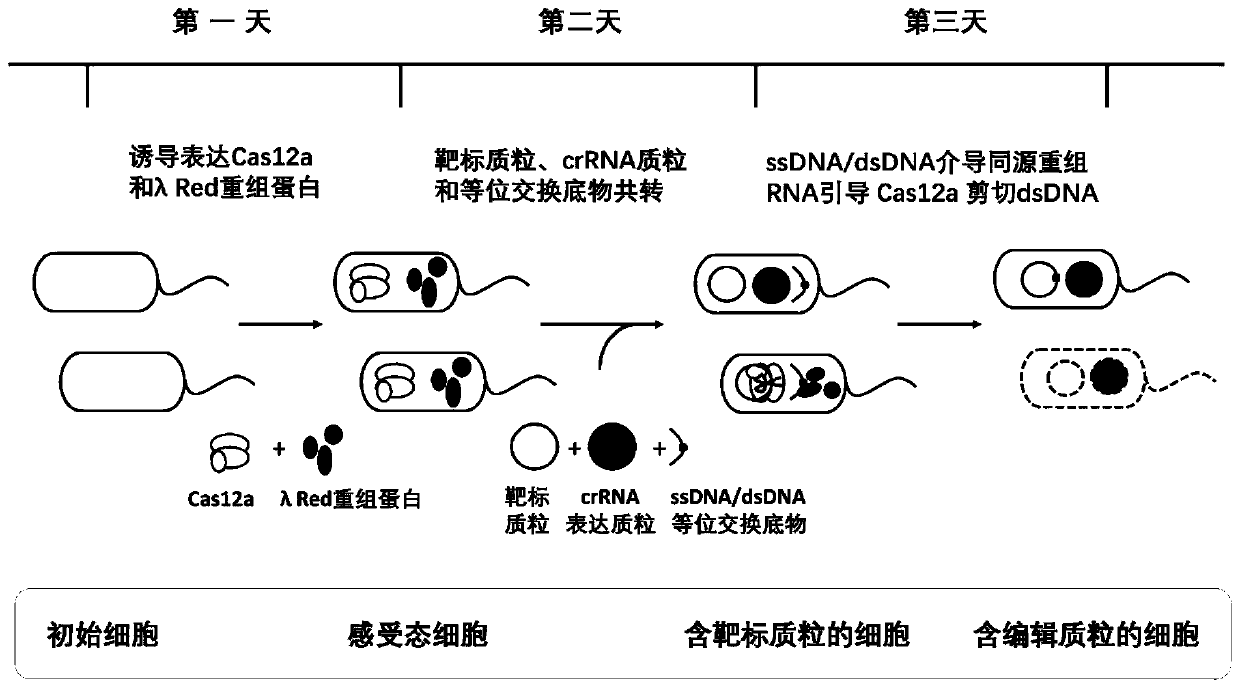
[0057] 4. Preparation of Competent Cells
[0058] i. Preparation of competent electroporation: select a single clone and culture it in 4mL culture medium at 30°C for 16-18h. 1:25 was transferred to the saturated bacterial solution, and the inducer 20% arabinose was added when the OD was 0.2. Continue to cultivate to 0.4-0.5, heat shock in a water bath shaker at 42°C for 15 minutes, and immediately place it on ice for 10-15 minutes. Centrifuge at 6500g for 7 minutes at 4°C to collect the cells. ddH 2 O Wash the cells twice, and finally resuspend the cells in 1 / 100 of the initial culture volume in pre-cooled 10% glycerol, and then pack into 50 μL / EP tubes for use.
[0059] ii. Preparation of Transformation Competent: Conventional CaCl 2 method to prepare the competent state.
[0060] 5. Transformation and screening of recombinants
[0061] 50ng of target plasmid, 200ng of crRNA expression plasmid and 1μg of ssDNA (or 700ng of dsDNA) were mixed, placed on ice, and then tran...
Embodiment 1
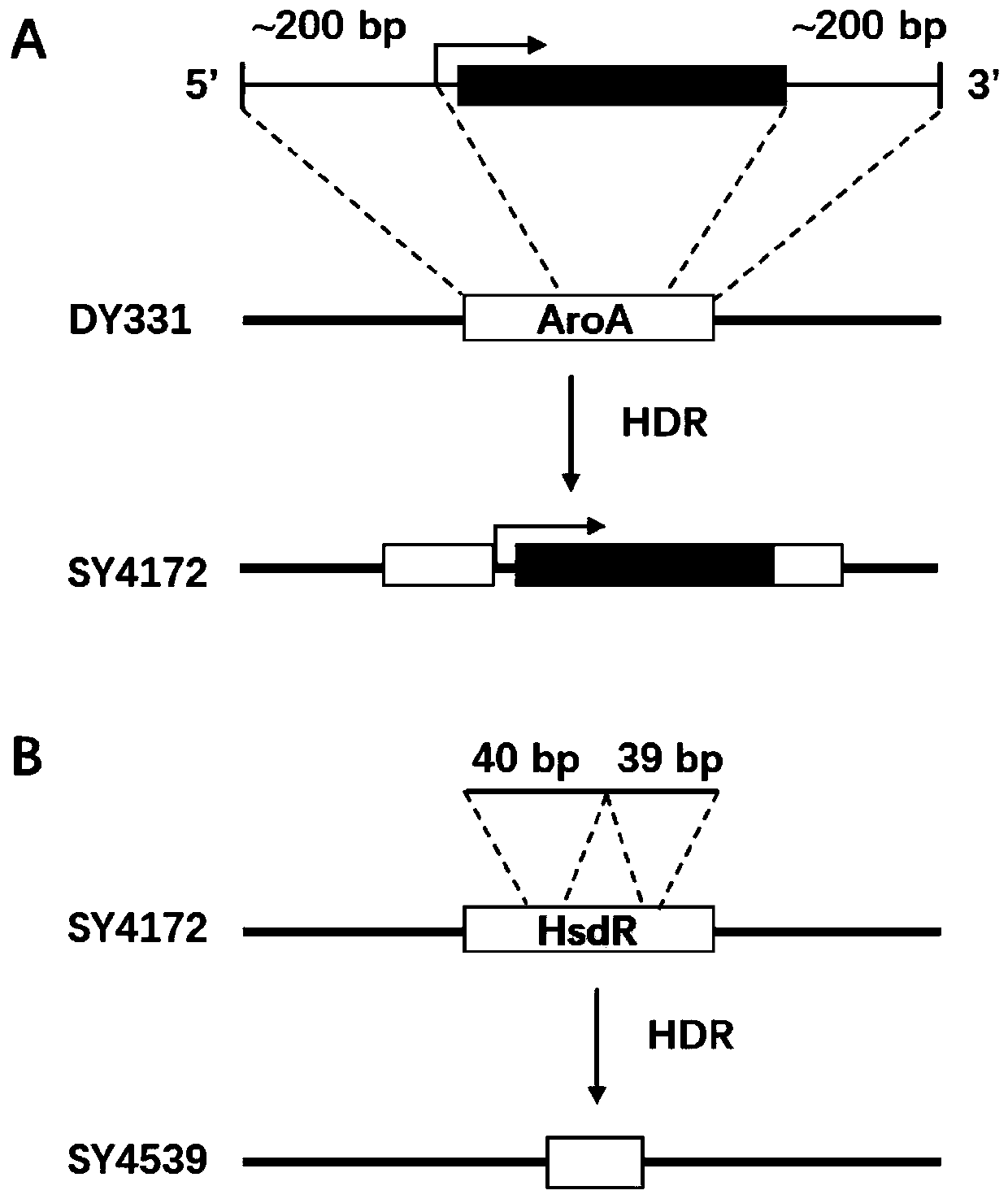
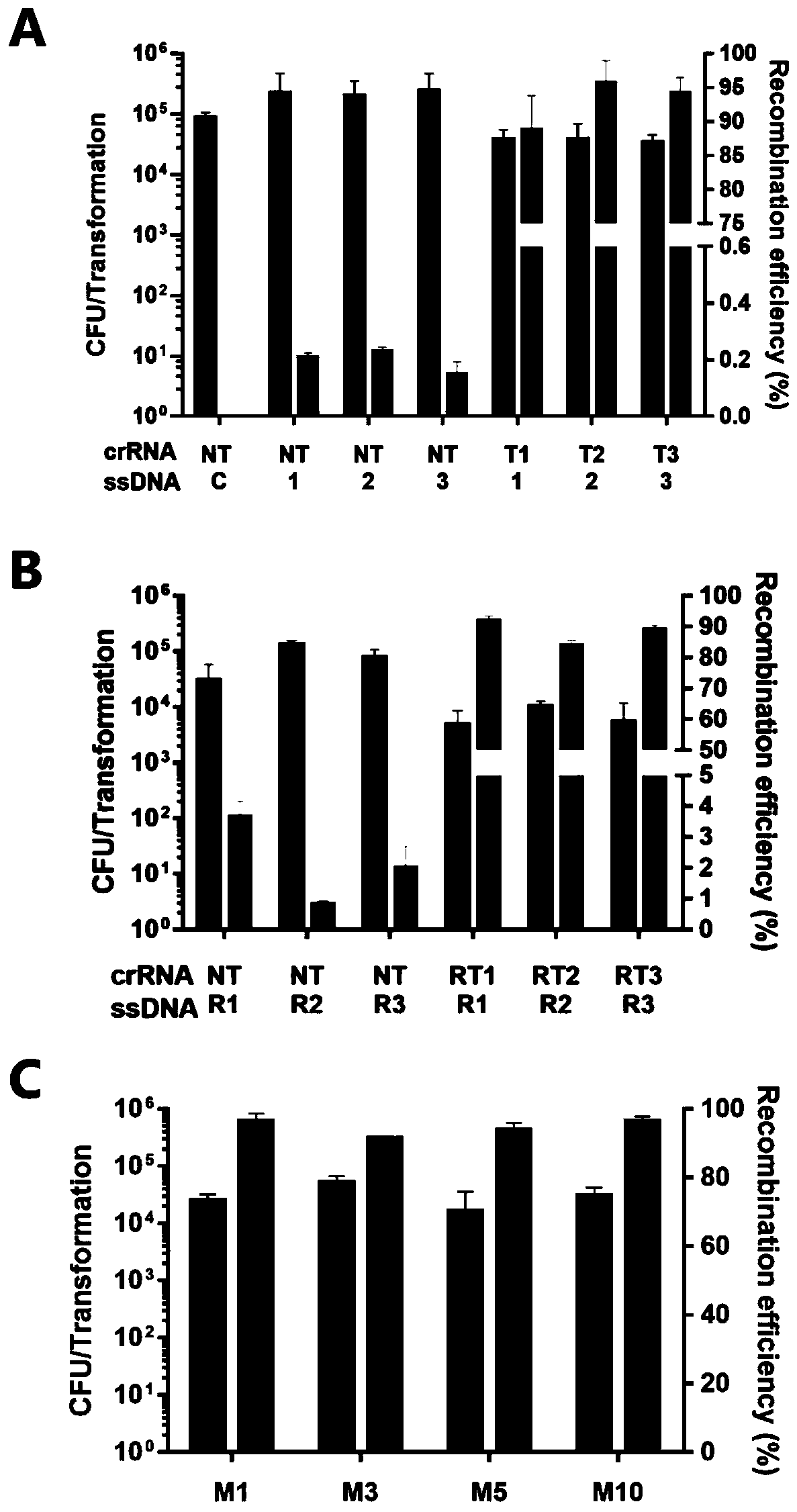
[0066] Embodiment 1. Fragment replacement of plasmid pJV53-GFP
[0067] 1) Find the crRNA action sites of the GFP-encoded gene on the plasmid pJV53-GFP, the sequences are T1, T2, T3; for each site, synthesize forward and reverse oligos, the sequences of which are shown in Table 1 as SEQ ID No: 1 and 2, 4 and 5, 7 and 8. Forward and reverse oligos were phosphorylated with T4 polynucleotide kinase (NEB) (37°C, 30min) and then annealed (95°C, 5min / °C and slowly cooled to 25°C after 5min), ligated into pAC- crRNA vector, construct the corresponding crRNA expression plasmid.
[0068] 2) Allelic exchange substrates oligo 1, 2, 3 designed for replacement by homologous recombination, the sequences of which are listed in Table 1 as SEQ ID No: 3, 6, 9.
[0069] 3) Preparation of SY4539 electroporation competent cells.
[0070] 4) Transfer the above-mentioned crRNA expression plasmid, original target plasmid pJV53-GFP and allelic exchange substrate oligo into the competent cells prepa...
Embodiment 2
[0073] Embodiment 2. Fragment replacement of plasmid pJV53-GFPm back mutation
[0074] 1) For the above-mentioned plasmid pJV53-GFPm (the three sites correspond to plasmids pJV53-GFPm1, pJV53-GFPm2 and pJV53-GFPm3 respectively) that have been successfully replaced through verification, search for crRNA action sites, the sequences of which are RT1, RT2, RT3; For the forward and reverse oligos of each site, the sequences thereof are shown in Table 1 as SEQ ID Nos: 10 and 11, 13 and 14, and 16 and 17. Forward and reverse oligos were phosphorylated by T4 polynucleotide kinase (NEB) (37°C, 30min) and then annealed (95°C, 5min / °C after 5min, slowly cooled to 25°C), and then ligated into pAC- crRNA vector, construct the corresponding crRNA expression plasmid.
[0075] 2) Allelic exchange substrates oligo R1, R2, R3 designed for homologous recombination to achieve replacement, the sequences are shown in SEQ ID No: 12, 15, 18 in Table 1.
[0076]3) Preparation of SY4539 electroporati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com