Kit for detection of human PIK3CA gene mutation and clinical application
A kit and base sequence technology, applied in the field of genetic engineering, can solve the problems of non-luminescence of probes and inability of instruments to detect fluorescent signals, and achieve the effects of scale, low cost, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1PI
[0056] Embodiment 1 PIK3CA wild-type plasmid, mutant plasmid, internal control plasmid construction
[0057] The vector used for the construction of the PIK3CA mutant plasmid is Puc57, with a full length of 2710 bp, constructed by Nanjing GenScript Biotechnology Co., Ltd. The wild-type plasmid fragment 400bp (NM_006218.2), the mutant plasmid fragment 400bp (NM_006218.2), and the internal control plasmid fragment 400bp (NM_006218.2) (the conserved region was selected as the internal control) were respectively connected with the vector to synthesize the plasmid.
Embodiment 2
[0058] The design of embodiment 2 primers, probes
[0059] According to the mutation information, Primer Premier 5.0 software was used to design primers and probes. At the same time, primers and probes were designed on exon 1 of PIK3CA as internal and external controls: the sequences of internal and external controls were consistent, and the modified reporter fluorescent groups were different.
[0060] The primer length is about 19-30 bases, the GC content is 40-60%, the Tm value is 59-61°C, and the amplified fragment is about 150bp. The missense mutation site is located at the last position of the 3' end of the upstream primer, and at the same time, a mismatch mutation base is introduced at different positions of the upstream primer, and the mutant upstream primer that meets the requirements is selected after screening:
[0061] G1624A:
[0062] F1: AAGCAATTTCTACACGAGATCCTCTCTCTA
[0063] F2: AAGCAATTTCTACACGAGATCCTCTCTCCA
[0064] F3: AAGCAATTTCTACACGAGATCCTCTCTCGA
[00...
Embodiment 3
[0111] The preliminary screening of embodiment 3 primers, probes
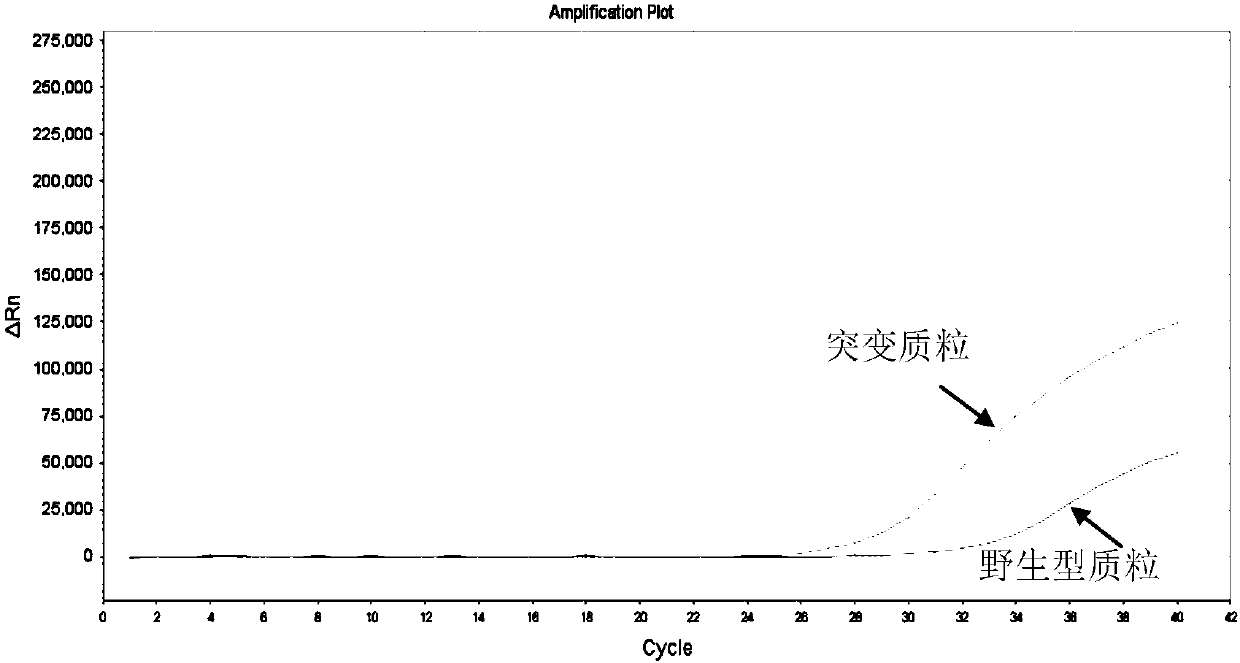
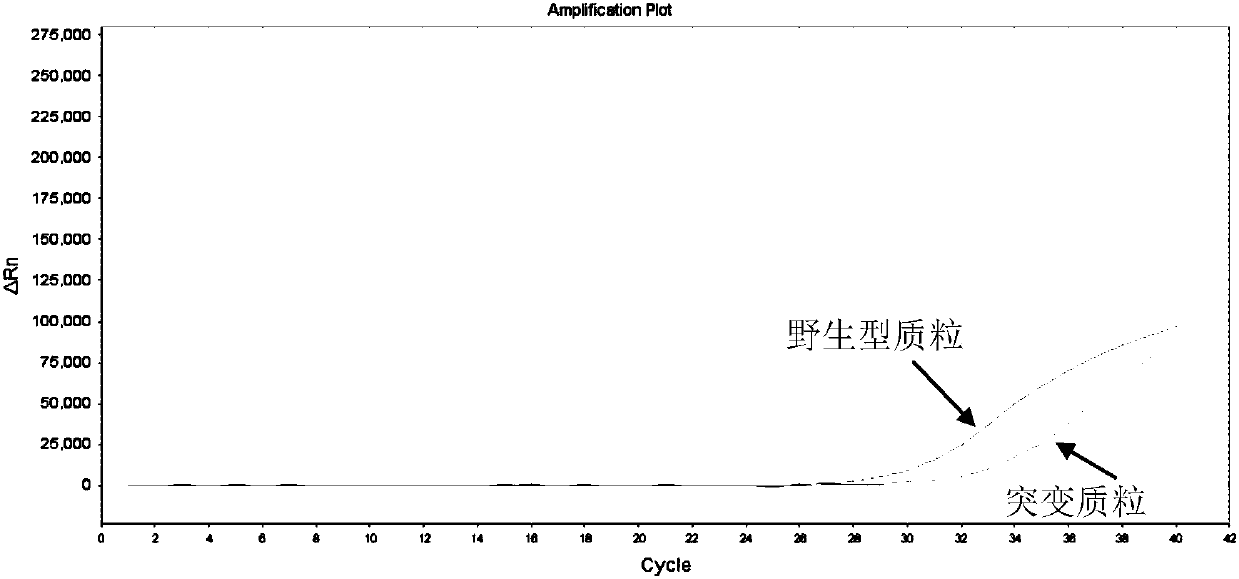
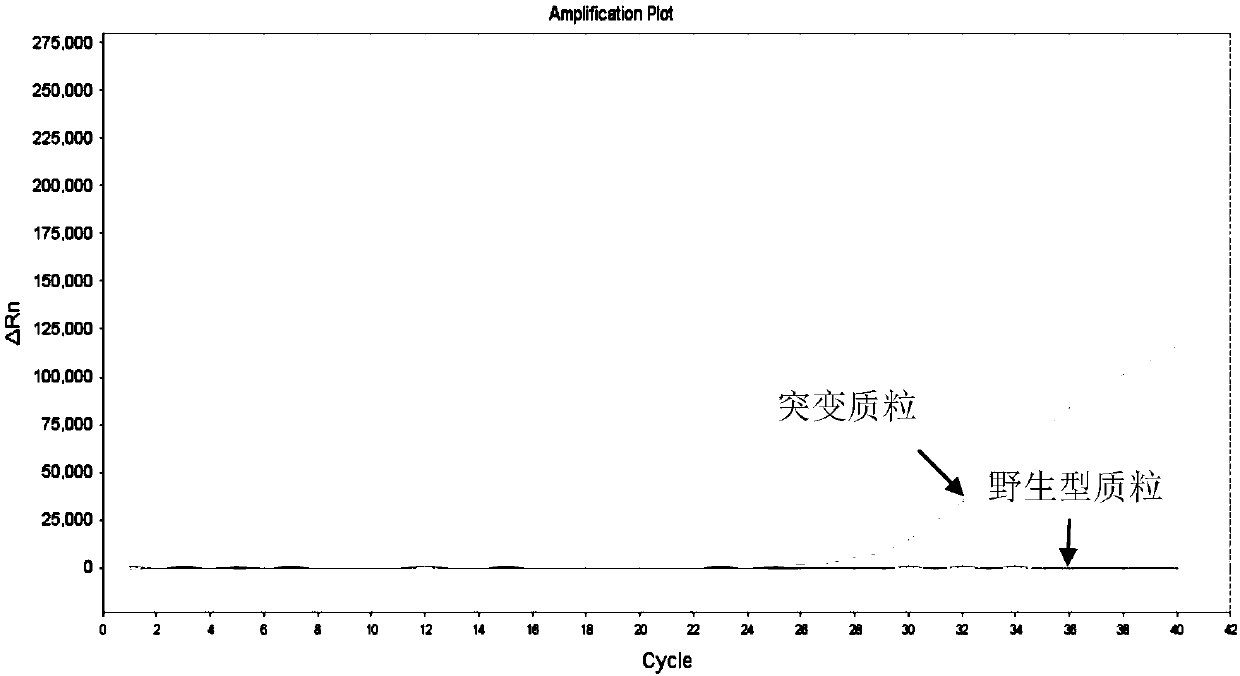
[0112] Amplify the mutant plasmid, the wild-type plasmid, and the internal control plasmid for the primer probes respectively, and the combination of the mutant primer probes that meets the requirements is: amplifying the mutant plasmid, and having no amplification reaction with the wild-type and internal control plasmids.
[0113] Table 2 Primer probe primary screening PCR reaction system
[0114]
[0115] PCR reaction program: 50°C for 10min; 95°C for 5min; 40 cycles of 95°C for 15sec and 60°C for 1min, and collect fluorescence at 60°C.
[0116] Wherein: the internal control / external control amplification fragment is shown in SEQ ID NO: 1;
[0117] The wild-type amplified fragments of G1624A, G1633A, and G1635T are shown in SEQ ID NO: 2;
[0118] A3140G, A3140T wild-type amplified fragments are shown in SEQ ID NO: 3;
[0119] The G1624A mutant amplified fragment is shown in SEQ ID NO: 4;
[0120] The G...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com