Method for non-invasive antenatal determination of aneuploidy by targeting next generation sequencing with biallelic SNP
A biallelic, aneuploidy technique for non-invasive prenatal testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0120] In one embodiment, an amplicon-based library preparation method is used, ie, target-specific primers are used to selectively amplify and thus enrich for target sequences. In one embodiment, the amplicon-based method comprises the steps of:
[0121] 1. Free DNA extracted from end repair and A-tail processing;
[0122] 2. Ligation of cell-free DNA with platform-specific adapter sequences, index sequences, and unique molecular identifiers (UMI);
[0123] 3. Wash away unconnected debris;
[0124] 4. Amplification of ligated free DNA using target-specific primers;
[0125] 5. Amplify the enriched fragments using universal primers; and
[0126] 6. Purification of the amplified target sequence.
[0127] In one embodiment, the index sequence is added to the target sequence in a post-enrichment amplification and ligation step.
[0128] As used herein, libraries are prepared based on amplicons, which are short synthetic DNA fragments that allow DNA ligated to the adapters to...
example 1
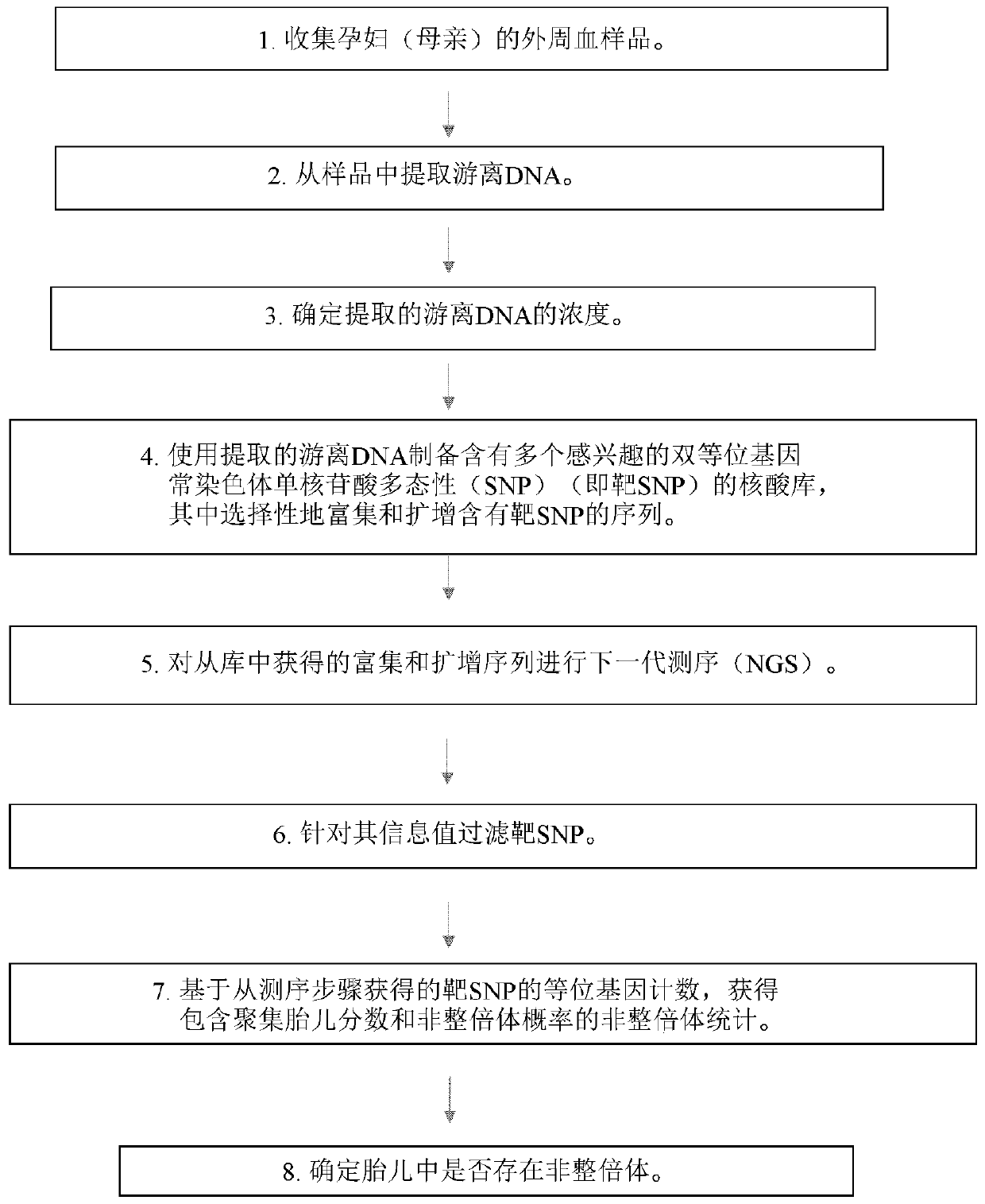
[0331] Sample Preparation - Purification of Cell-Free DNA from Maternal Blood or Plasma Samples
[0332] This example illustrates the use of a The Rapid Sample Concentrator (RSC) extracts cell-free DNA from maternal whole blood or plasma samples and uses Example of Fluorimeter Measurement of Extracted Genomic DNA Concentration.
[0333] 10 mL whole blood samples were collected from pregnant subjects and stored in cfDNA blood tubes. Samples were then processed according to the following scheme:
[0334] Prepare plasma
[0335] 1. Centrifuge whole blood from cfDNA blood tubes at 3000 rpm for 5 minutes.
[0336] 2. Aliquot all plasma and centrifuge collected plasma at 14000 rpm for 10 minutes.
[0337] 3. Collect the supernatant in a new 2 mL screw cap tube and store at 4°C until further use, or -20°C for long-term storage.
[0338] Binding of circulating nucleic acids to magnetic resins
[0339] 1. Add 2mL of Binding Buffer to a 50mL centrifuge tube.
[0340] 2. Ad...
example 2
[0384] Hybridization-Based Targeted Enrichment-Library Preparation Protocol
[0385] In the present invention, hybridization-based library preparation was built on the SeqCap manufacturer's protocol. In the following protocols and procedures, minor differences from the manufacturer's version were introduced to customize and optimize the workflow for better sequence reads at subsequent stages.
[0386] The process includes DNA fragmentation, end repair and A-tail processing, adapter ligation, library amplification (i.e., pre-enrichment amplification), post-amplification cleanup, and sample hybridization with the SeqCap probe pool (i.e., target enrichment amplification). ) and post-hybridization amplification (i.e. post-enrichment amplification). The detailed plan is as follows:
[0387] A. Bring AMPure XP beads to room temperature
[0388] B. Prepare DNA (free DNA is recommended to use 1ng-1μg)
[0389] 1. Maximize volume - 50 µL cfDNA input
[0390] C. End repair and A-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com