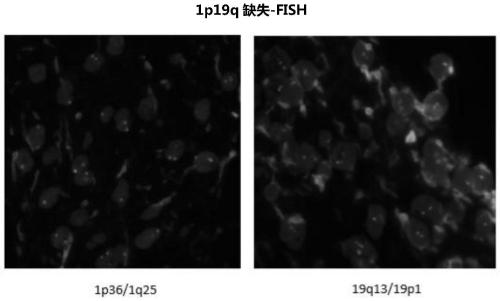
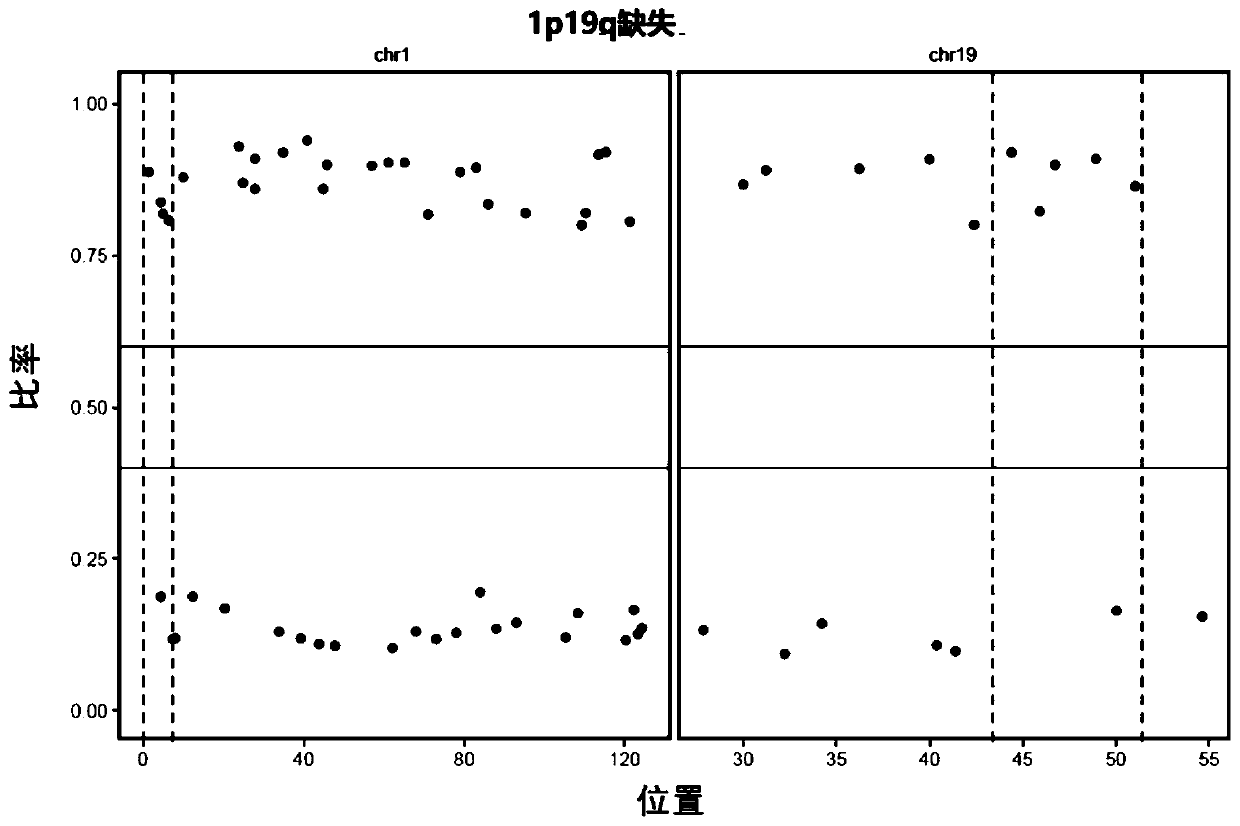
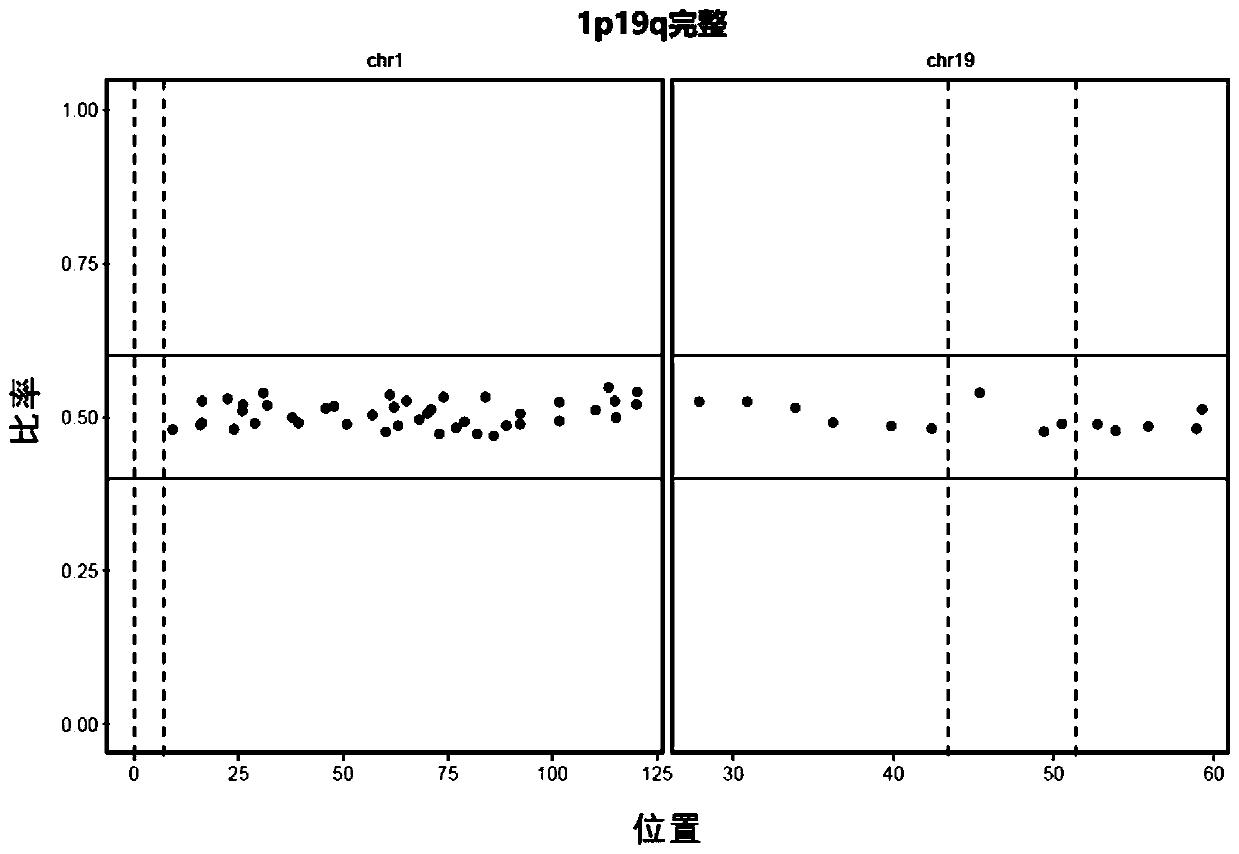
Neuroglioma 1p/19q co-deletion detection system based on next generation sequencing
A glioma and next-generation sequencing technology, applied in the system field of glioma 1p/19q joint deletion detection, can solve the problems of reducing the accuracy of the results, inconvenient, accurate judgment of the results, and complicated operations, etc., to achieve improved Assay Accuracy, Accuracy Combined Effect of Deletion Identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0054] According to a typical embodiment of the present invention, the system includes a first verification device, the first verification device is used for STR-based joint deletion detection of 1p and 19q, and the first verification device includes: a STR acquisition module, used for obtaining from existing data Extract known STR; control sample STR statistics module, used to extract the sequencing sequence near the known STR from the comparison result file of the control sample, count the number of repetitions of the known STR on each read, according to the degree of coverage of the read on the STR and The sequencing coverage of the STR region, count the number of reads of each STR repeat, extract the two STR repeats with the largest number of reads, and record it as N 3 and N 4 ;like Then the STR is considered to be homozygous and is no longer used for result judgment; preferably, n>5; more preferably, n=10. The STR statistics module of the sample to be tested is used t...
Embodiment 1
[0106] 1. Sample preparation: fragmentation and capture steps of tissue sample DNA
[0107] (1), tissue DNA extraction and interruption:
[0108] Extract tissue DNA using a tissue extraction kit. The extracted DNA was quantified using Qubit 3.0 and dsDNA HS Assay Kit.
[0109] Cut the polytetrafluoroethylene thread to a length of about 1 cm with medical scissors sterilized by ultraviolet light, and ensure that the length of the breaking rod is uniform. Place it in a clean container and sterilize it by ultraviolet light for 3 to 4 hours. After the sterilization is completed, put the 1cm polytetrafluoroethylene line into the 96-well plate with sterilized tweezers. Put 2 break rods in each well, and then sterilize the 96-well plate by ultraviolet for 3 to 4 hours after completion.
[0110] Take 300ng tissue DNA samples according to the qubit quantitative results, use TE to dilute to 50 μl, transfer to a 96-well plate, place the tin foil membrane on the 96-well plate, align the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com