Double-fluorescence screening method for fungal gene knockout
A gene knockout and screening method technology, applied in the field of fungal gene knockout, can solve the problems of distinguishing difficult-to-transformants, etc., and achieve the effect of omitting the verification process and reducing the workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0073] Hereinafter, the present invention will be described with reference to examples, but the present invention is not limited to the following examples.
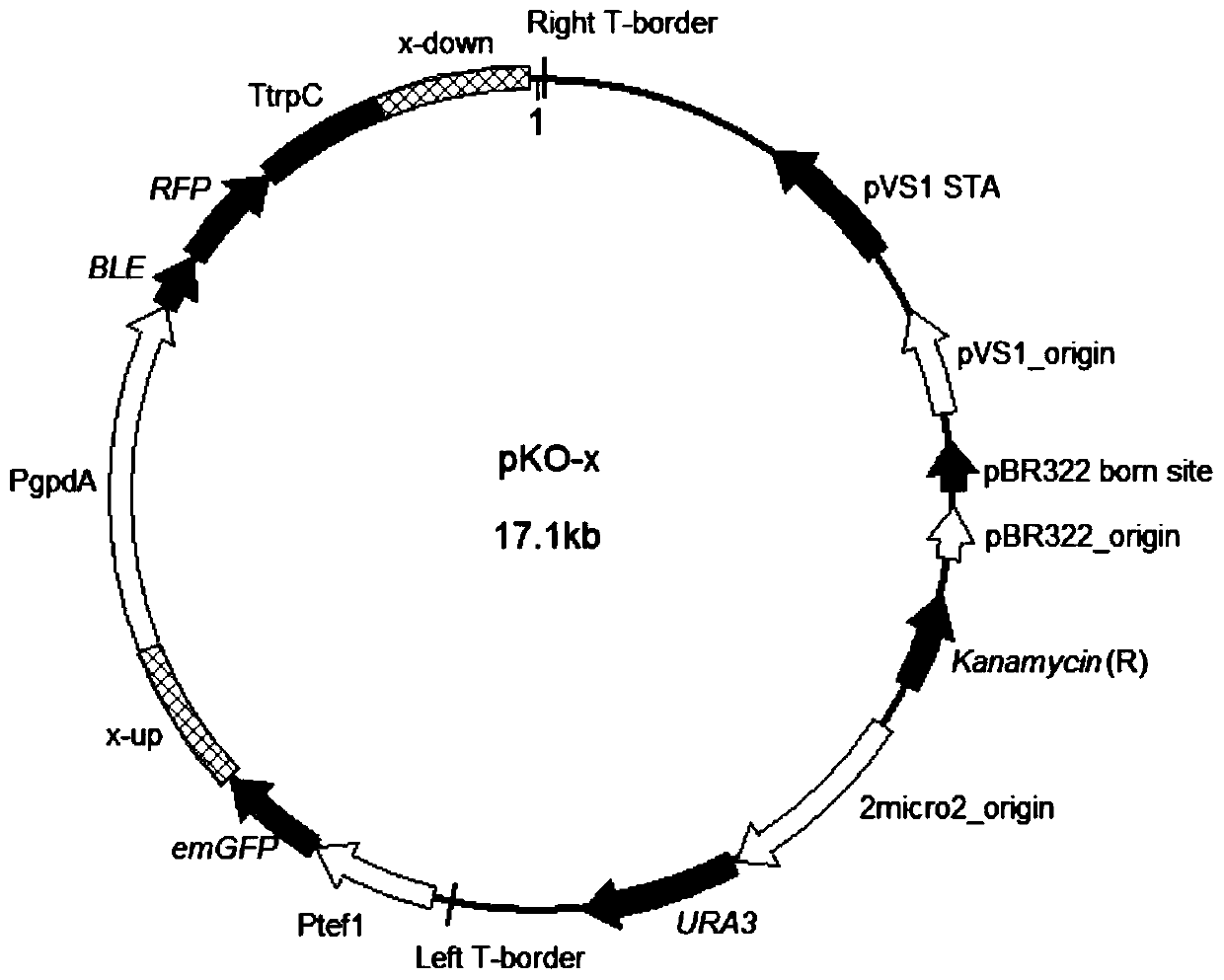
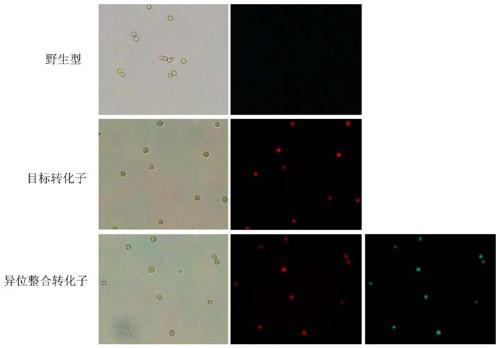
[0074] This embodiment is applicable to fungi sensitive to bleomycin. Aspergillus flavus NRRL 3357 is a whole-genome sequenced bacterium and is sensitive to bleomycin, so Aspergillus flavus is selected as the implementation object.
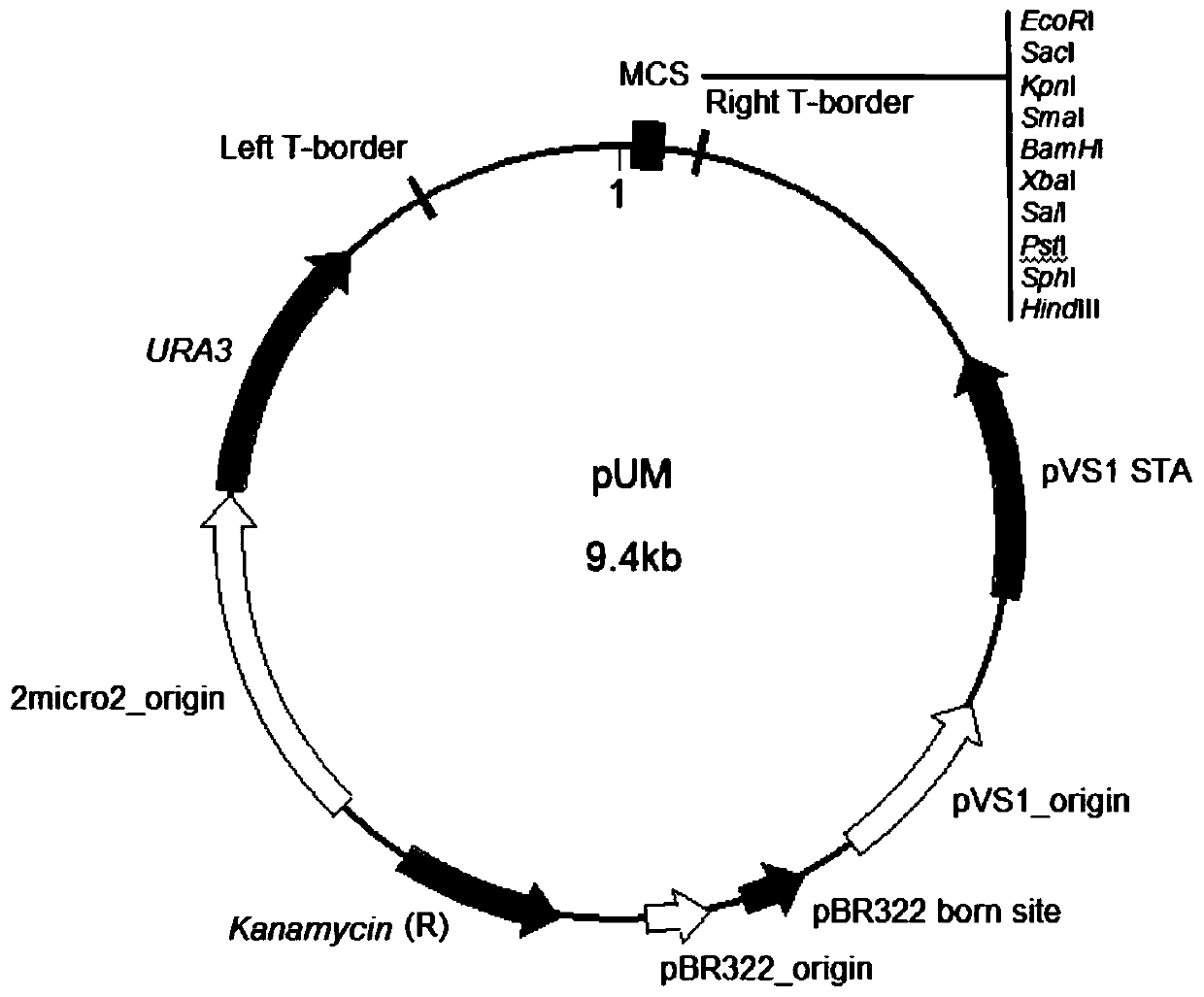
[0075] pUM vector construction
[0076] 1) Using the pYES2 vector DNA as a template, use Primerdesign software to design primers:
[0077] Primer ① ccgcggGGAACAACACTCAACCCTA;
[0078] Primer ② ccgcggTTCGATGTAACCCACTCG;
[0079] The lowercase letter part in the above primers is the restriction enzyme cutting site SacII.
[0080] 2) Using the above primers ① and ②, amplify the 2.9-kb URA3-2micro2_origin fragment from the template in step 1), and then insert pCAMBIA1300 into the SacII site of the backbone vector. The constructed vector was named pUM.
[0081] Among them, the amplification...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com