Maize Molecular Breeding Core SNP Site Combination and Its Application
A maize and site technology, which is applied to the combination of core SNP sites in maize molecular breeding and its application field, can solve the problems of application limitation, site fixation and high detection cost, and achieve the effect of improving breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0365] Example 1, Screening of Molecular Breeding Core SNP Site Combinations and Design of KASP Primers
[0366] 1. Screening of core SNP loci in molecular breeding
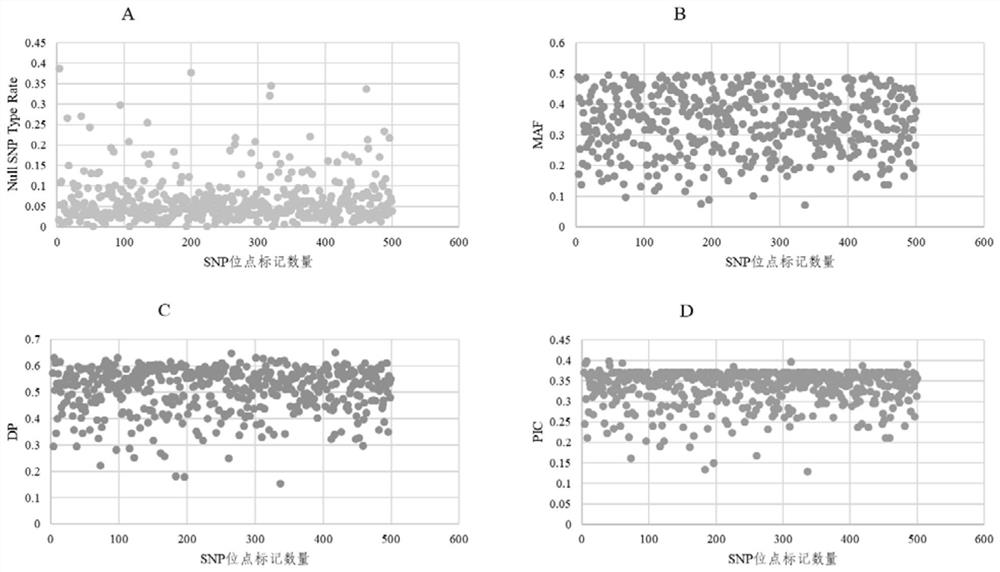
[0367] 1. Preliminary screening of 740 SNP sites
[0368] From the Maize6H90K chip developed by the Maize Research Center of Beijing Academy of Agriculture and Forestry Sciences, 740 SNP sites were selected according to the comprehensive indicators such as site typing effect, chromosome uniform distribution, PIC value, and MAF value, and 740 sets of KASP primers were designed and synthesized. A primer corresponds to a SNP site.
[0369] 2. Re-screen 498 pairs of SNP markers with ideal typing effect
[0370] The genomic DNA of 157 maize materials to be identified was extracted and diluted to 200ng / μl (RNAase enzyme treatment) respectively, and prepared into two 96-well mother liquid plates, and the remaining sample wells were used as reference NTC.
[0371] A total of 740 pairs of primers were synthesized to pe...
Embodiment 2
[0705] Example 2, Molecular Breeding Core SNP Sites and Application of KASP Primers
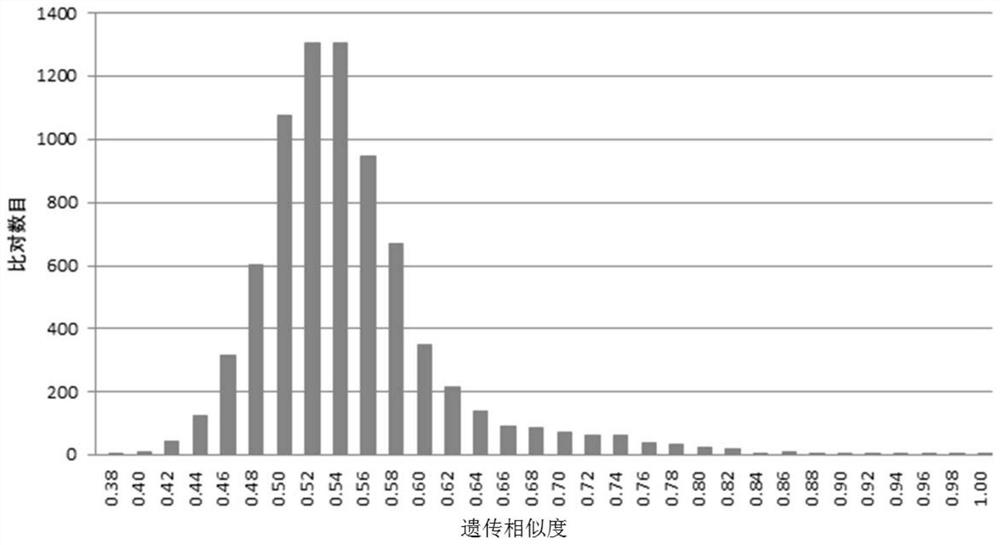
[0706] 1. The application of molecular breeding core SNP loci and their KASP primers in detecting the genetic similarity of inbred lines and detecting the heterozygosity of hybrids
[0707] 1. Detection of genetic similarity of inbred lines
[0708] Genomic DNA was extracted from the 124 corn inbred lines shown in Table 1 as templates, respectively amplified with the KASP primers shown in Table 3, and the genotypes of 300 core SNP loci of the 124 corn inbred lines were detected.
[0709] The brief workflow of the KASP screening primer test is as follows: Dilute the DNA from the mother plate to 20ng / μl as the Master plate; use the Replikator hole plate duplicator of LGC Company to select the appropriate mode to divide the liquid into the 1536-well Working plate, and Import the liquid separation data into the Kraken system; use the drying oven to dry the DNA in the Working plate; use the Merdi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com