A method for constructing a single-cell RNA sequencing library
A sequencing library and construction method technology, applied in the field of single-cell RNA sequencing, can solve the unsatisfactory problems such as the inability to obtain the full-length transcript chain information of multiple transcript RNAs, and the untrue expression of gene expression, so as to avoid expansion The effect of increasing bias
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
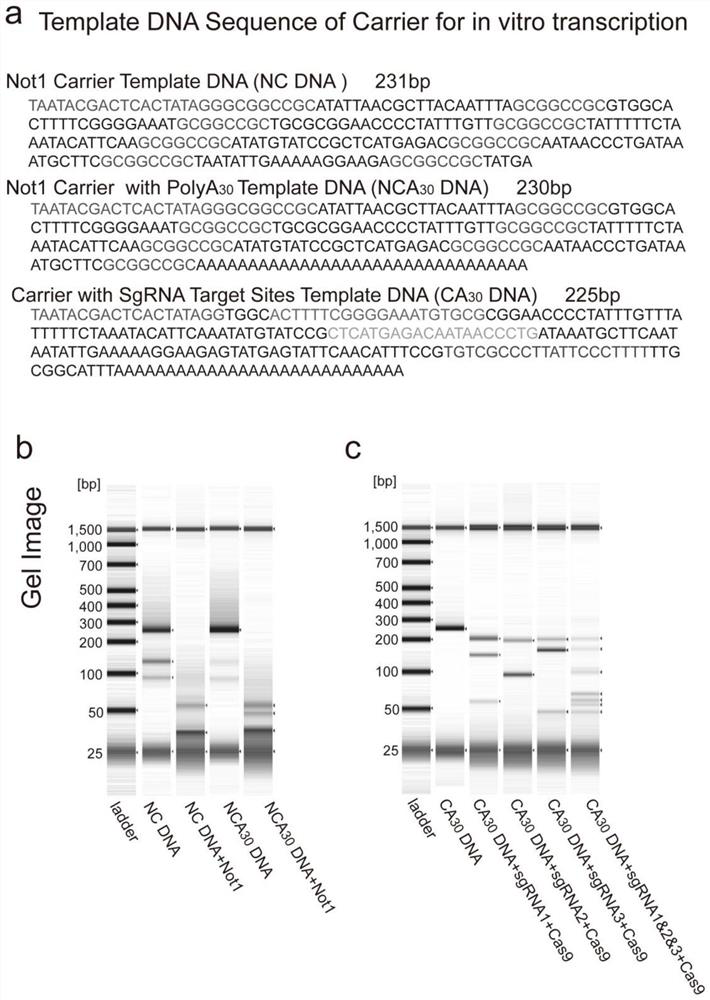
[0063] Preparation of carrier RNA
[0064] The corresponding DNA fragments of the carrier RNA containing the 3' end poly A tail and the carrier RNA sequence not containing the 3' end poly A tail can add T7 promoter sequence at the 5' end (Schenborn et al., 1985 ) into the carrier (bacterial plasmid), the complete DNA sequence containing the T7 promoter can be double-digested through the restriction enzyme site on the carrier, and the DNA fragments are separated by agarose gel electrophoresis, and the gel is recovered to obtain the target fragment. The target DNA fragment obtained after purification was obtained by in vitro transcription kit (HiScribe T7 Quick High Yield RNA Synthesis Kit, NEB#2050) to obtain carrier RNA, carrier RNA larger than 200 bases can be obtained by PlusMini Kit (Qiagen) for purification, the carrier RNA less than 200 bases can be purified by MEGAclear TM KitPurification for Large Scale Transcription Reactions (Ambion) for purification, smallcarrier...
Embodiment 1
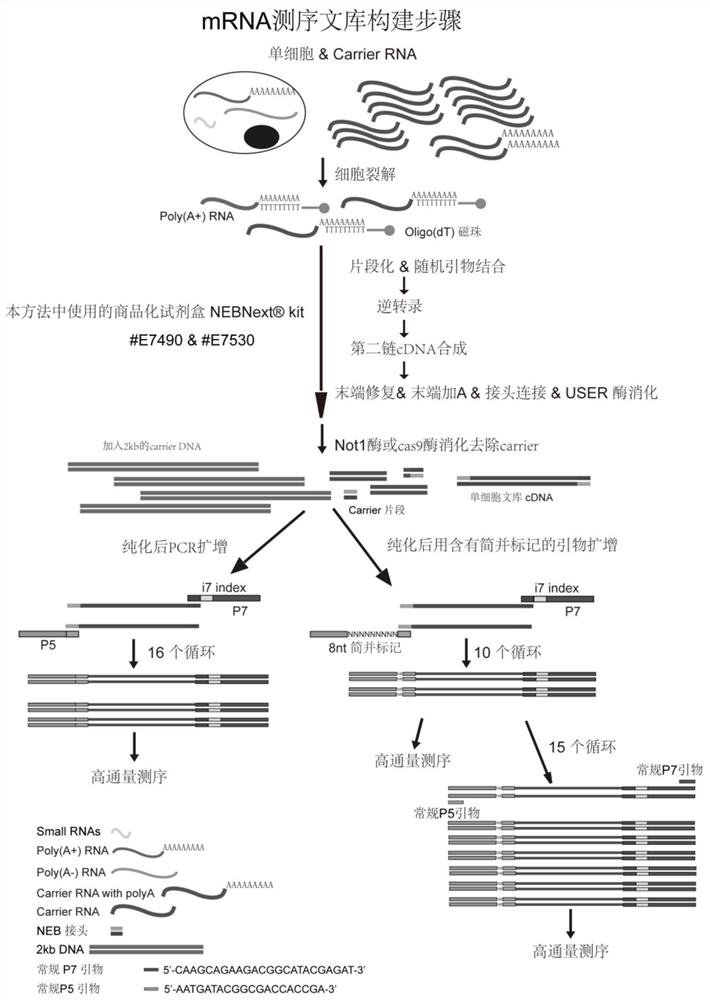
[0066] Example 1 Poly (A) selected single-cell RNA library construction (see the attached flowchart for details figure 2 )
[0067] 1.1 Obtain single cells by micromanipulation or flow cytometry for lysis. The method of lysis can be heated at 95°C for 5 minutes in 0.1% BSA / PBS or other non-ionic surfactants. Add about 100ng of carrier RNA, the carrier RNA here is a mixed sample, 5%-10% of which is a carrier RNA (SEQ ID NO.:1) containing a 3'-end polyadenylic acid tail, and the rest is a carrier RNA that does not contain a 3'-end polyadenylic acid tail Adenylate-tailed carrier RNA (SEQID NO.:2), the final volume is 25 μL, and the insufficient part can be made up with nuclease-free water.
[0068] The next process of building a library starts with Ultra TM RNA Library Prep Kit for Illumina (#E7530) kit method as an example:
[0069] 1.2 by NEBNext Oligo d(T) 25 10 μL of magnetic beads to obtain RNA containing Poly(A), Poly(A) mRNAMagnetic Isolation Module (NEB#E7490)
[...
Embodiment 2
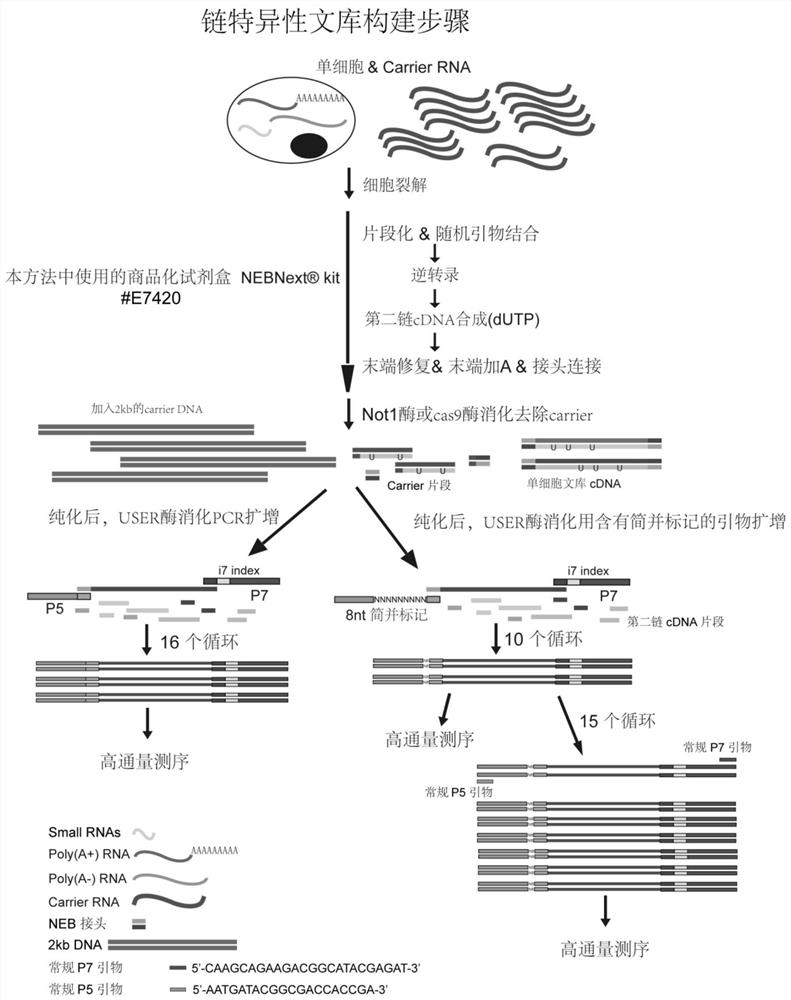
[0117] Example 2 Non-Poly(A)-selected chain-specific single-cell RNA library construction (see the attached flowchart for details image 3 )
[0118] 2.1 Obtain single cells by micromanipulation or flow cytometry for lysis. The method of lysis is to heat at 95°C for 5 minutes in 0.1% BSA / PBS or a lysate prepared with other non-ionic surfactants, and add about 100ng of carrier at the same time RNA, the carrier RNA here is the carrier RNA (SEQ ID NO.: 2) that does not contain the 3'-end polyadenylic acid tail, the final system is 3 μL, and the insufficient part can be made up with nuclease-free water.
[0119] The next process of building a library starts with Ultra TM Directional RNA Library Prep Kit for (#E7420) kit method as an example:
[0120] 2.2 Fragmentation of RNA and random primer binding
[0121]
[0122] Mix well, with a total volume of 12 μL, 94°C, 15 minutes, fragment the RNA to a size of about 200 bases (the adjustment of the fragment size can change th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com